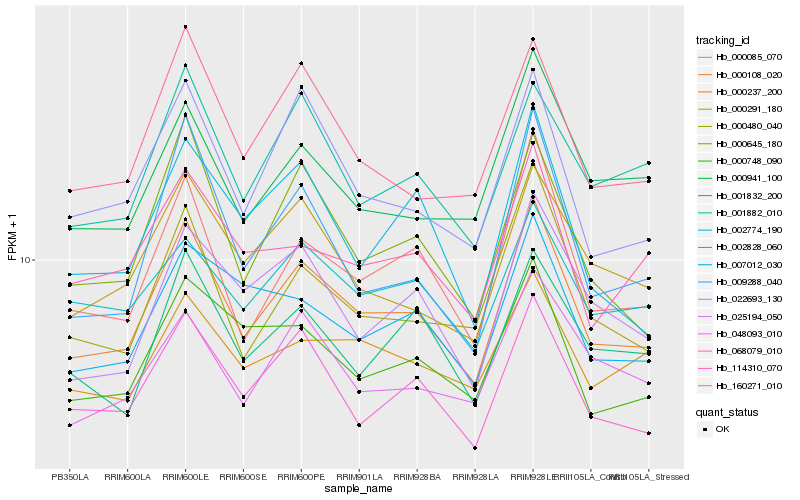
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000108_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 2 |
Hb_068079_010 |
0.0656104257 |
- |
- |
- |
| 3 |
Hb_000085_070 |
0.0687771377 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_022693_130 |
0.0768826631 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_160271_010 |
0.077087745 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_000645_180 |
0.0783307757 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 7 |
Hb_000480_040 |
0.0790616678 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 8 |
Hb_000291_180 |
0.0803144414 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 9 |
Hb_025194_050 |
0.0832143456 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001882_010 |
0.0842834575 |
- |
- |
- |
| 11 |
Hb_000748_090 |
0.0848262588 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 12 |
Hb_001832_200 |
0.0859207163 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007012_030 |
0.0868104373 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_048093_010 |
0.0874015459 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_000941_100 |
0.0881347313 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 16 |
Hb_002774_190 |
0.0884594508 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 17 |
Hb_114310_070 |
0.0886109184 |
- |
- |
GTP-dependent nucleic acid-binding protein engD, putative [Ricinus communis] |
| 18 |
Hb_009288_040 |
0.0910258129 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 19 |
Hb_000237_200 |
0.0919094399 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 20 |
Hb_002828_060 |
0.0926055558 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |