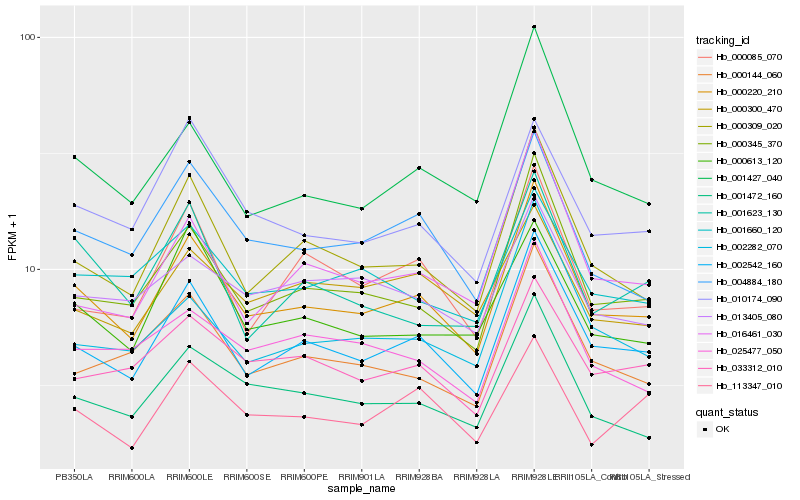
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000220_210 |
0.0 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002542_160 |
0.0412670961 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000309_020 |
0.0636042017 |
- |
- |
PREDICTED: uncharacterized protein LOC105641764 [Jatropha curcas] |
| 4 |
Hb_000345_370 |
0.0647875512 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P [Jatropha curcas] |
| 5 |
Hb_001427_040 |
0.0769550167 |
- |
- |
PREDICTED: uncharacterized protein LOC105637784 [Jatropha curcas] |
| 6 |
Hb_001472_160 |
0.0774601375 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 7 |
Hb_004884_180 |
0.0795316455 |
- |
- |
PREDICTED: protein TIC 56, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000144_060 |
0.085668365 |
- |
- |
PREDICTED: uncharacterized protein At5g03900, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_002282_070 |
0.0891918647 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |
| 10 |
Hb_033312_010 |
0.0898959402 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_025477_050 |
0.091305417 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_000613_120 |
0.0915784833 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000085_070 |
0.0929097264 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_010174_090 |
0.093584908 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_016461_030 |
0.0960122104 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_001660_120 |
0.0971028657 |
- |
- |
PREDICTED: adenylate kinase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_013405_080 |
0.0981827035 |
- |
- |
PREDICTED: proline--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_001623_130 |
0.1010486336 |
- |
- |
PREDICTED: GDT1-like protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_113347_010 |
0.1011689781 |
- |
- |
hypothetical protein CISIN_1g009336mg [Citrus sinensis] |
| 20 |
Hb_000300_470 |
0.1015511504 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |