| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
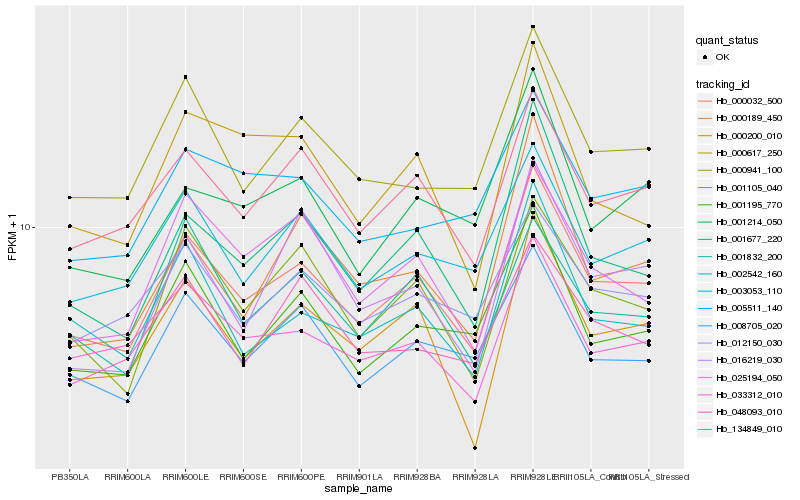
Hb_000032_500 |
0.0 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 2 |
Hb_134849_010 |
0.066061713 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 3 |
Hb_001677_220 |
0.0697411449 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 4 |
Hb_001105_040 |
0.0751811043 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000941_100 |
0.0761408562 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 6 |
Hb_000200_010 |
0.0776082566 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 7 |
Hb_008705_020 |
0.0779964086 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_003053_110 |
0.078443031 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001195_770 |
0.0803080466 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 10 |
Hb_002542_160 |
0.0817391096 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_025194_050 |
0.0837740296 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 12 |
Hb_033312_010 |
0.0844539528 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_012150_030 |
0.0855898666 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001214_050 |
0.0876974281 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001832_200 |
0.0913817454 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_048093_010 |
0.0939975446 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000189_450 |
0.0947355954 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_005511_140 |
0.0948527965 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 19 |
Hb_000617_250 |
0.0951042492 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_016219_030 |
0.0956975794 |
- |
- |
cytochrome P450, putative [Ricinus communis] |