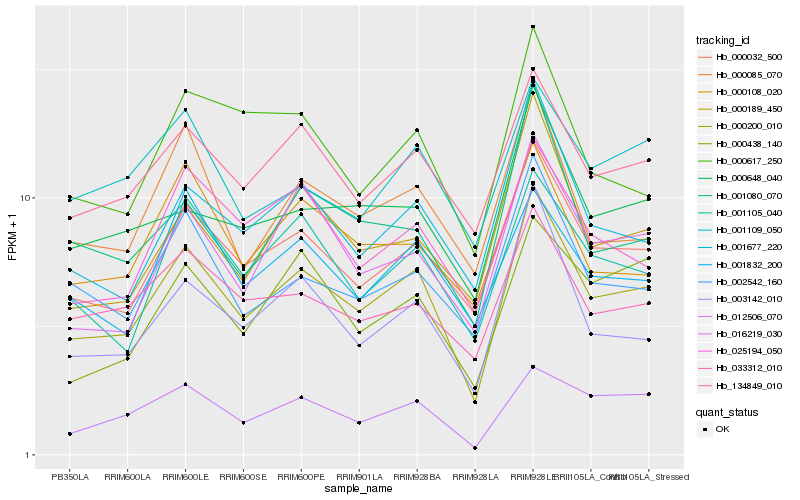
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000200_010 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein 8-like [Jatropha curcas] |
| 2 |
Hb_000032_500 |
0.0776082566 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 3 |
Hb_134849_010 |
0.0928403008 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 4 |
Hb_003142_010 |
0.093147752 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 5 |
Hb_001677_220 |
0.0979719809 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 6 |
Hb_001105_040 |
0.0998893239 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_012506_070 |
0.1004725484 |
- |
- |
hypothetical protein POPTR_0004s20060g [Populus trichocarpa] |
| 8 |
Hb_016219_030 |
0.1029972725 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_002542_160 |
0.1032443456 |
- |
- |
PREDICTED: peptide chain release factor APG3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000189_450 |
0.1059305389 |
- |
- |
Zeta-carotene desaturase, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000085_070 |
0.1064483404 |
- |
- |
PREDICTED: trigger factor-like protein TIG, Chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_033312_010 |
0.1070680177 |
- |
- |
PREDICTED: valine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_025194_050 |
0.1087626454 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001080_070 |
0.1093922298 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 15 |
Hb_000648_040 |
0.1099915228 |
- |
- |
unknown [Populus trichocarpa] |
| 16 |
Hb_000617_250 |
0.113240804 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000438_140 |
0.1145306605 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 18 |
Hb_000108_020 |
0.1153967828 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 19 |
Hb_001832_200 |
0.1158152336 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001109_050 |
0.1201640731 |
- |
- |
PREDICTED: ankyrin-1 [Jatropha curcas] |