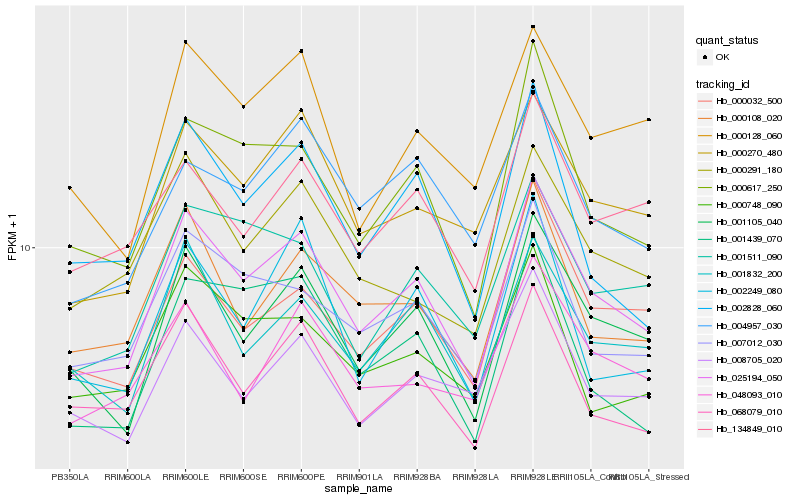
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025194_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001105_040 |
0.0617406156 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 10, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000617_250 |
0.0743932032 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000270_480 |
0.0772301078 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 5 |
Hb_001832_200 |
0.0811768449 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007012_030 |
0.0818855642 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 4-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_068079_010 |
0.0828562716 |
- |
- |
- |
| 8 |
Hb_000108_020 |
0.0832143456 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 9 |
Hb_000032_500 |
0.0837740296 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 10 |
Hb_001511_090 |
0.0883113324 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 11 |
Hb_000291_180 |
0.0909701173 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 12 |
Hb_134849_010 |
0.0914076762 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 13 |
Hb_004957_030 |
0.09170145 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 14 |
Hb_000128_060 |
0.0924379138 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 15 |
Hb_002828_060 |
0.0941076934 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001439_070 |
0.094463241 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 17 |
Hb_048093_010 |
0.0951823499 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_008705_020 |
0.0951921704 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_002249_080 |
0.0960117108 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 20 |
Hb_000748_090 |
0.096393057 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |