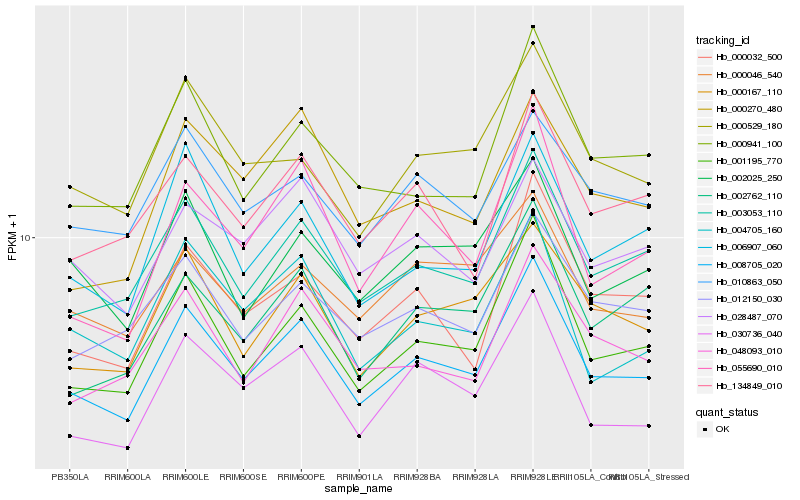
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008705_020 |
0.0 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_001195_770 |
0.0470348219 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 3 |
Hb_003053_110 |
0.055801408 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_030736_040 |
0.0682303033 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000167_110 |
0.0708126626 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_006907_060 |
0.0727684732 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_000941_100 |
0.0733915083 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 8 |
Hb_055690_010 |
0.0734404645 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_012150_030 |
0.0770781636 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002025_250 |
0.0771683358 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 3 [Jatropha curcas] |
| 11 |
Hb_000032_500 |
0.0779964086 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 12 |
Hb_004705_160 |
0.0784603488 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 13 |
Hb_000529_180 |
0.0789137188 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_048093_010 |
0.0795002163 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_002762_110 |
0.0814802973 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_134849_010 |
0.0817247268 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 17 |
Hb_000046_540 |
0.0817958381 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 18 |
Hb_028487_070 |
0.0821378893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010863_050 |
0.083829167 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 20 |
Hb_000270_480 |
0.0877890083 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |