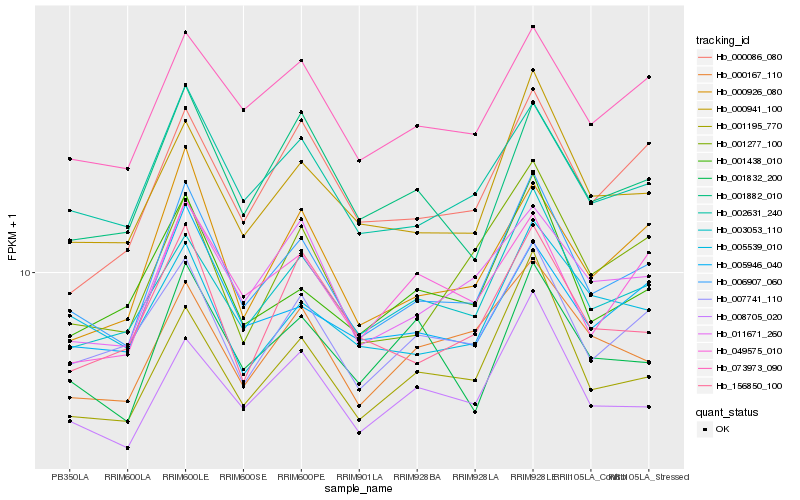
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006907_060 |
0.0 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_002631_240 |
0.0617916639 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 3 |
Hb_000941_100 |
0.0671682466 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 4 |
Hb_003053_110 |
0.068611617 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_073973_090 |
0.071183768 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 6 |
Hb_008705_020 |
0.0727684732 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 7 |
Hb_001195_770 |
0.0727837818 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 8 |
Hb_007741_110 |
0.0732889865 |
- |
- |
- |
| 9 |
Hb_011671_260 |
0.0762113162 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000926_080 |
0.0793266723 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 11 |
Hb_001438_010 |
0.0803970865 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001882_010 |
0.0839554452 |
- |
- |
- |
| 13 |
Hb_005946_040 |
0.084289607 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000086_080 |
0.0867654663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000167_110 |
0.0869039494 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_001277_100 |
0.08829897 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 17 |
Hb_049575_010 |
0.0889918282 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 18 |
Hb_156850_100 |
0.0893501695 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 19 |
Hb_001832_200 |
0.0897328229 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_005539_010 |
0.0898953562 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |