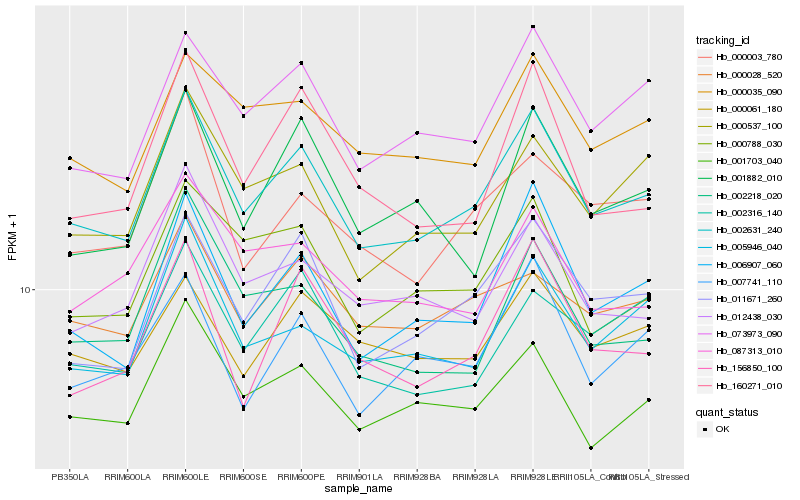
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002631_240 |
0.0 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 2 |
Hb_006907_060 |
0.0617916639 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_011671_260 |
0.0618764207 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_073973_090 |
0.063867513 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 5 |
Hb_001703_040 |
0.0658144702 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000537_100 |
0.0721838551 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_005946_040 |
0.0738617375 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000035_090 |
0.0749901117 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 9 |
Hb_000788_030 |
0.0769616419 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 10 |
Hb_000028_520 |
0.0781110294 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 11 |
Hb_000003_780 |
0.0791359858 |
- |
- |
hexokinase [Manihot esculenta] |
| 12 |
Hb_012438_030 |
0.0796446135 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 13 |
Hb_007741_110 |
0.0797764419 |
- |
- |
- |
| 14 |
Hb_000061_180 |
0.0798053289 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 15 |
Hb_156850_100 |
0.0813007638 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 16 |
Hb_160271_010 |
0.0828631164 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_001882_010 |
0.0831462544 |
- |
- |
- |
| 18 |
Hb_002218_020 |
0.0837601675 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 19 |
Hb_087313_010 |
0.0848257523 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002316_140 |
0.0862287094 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |