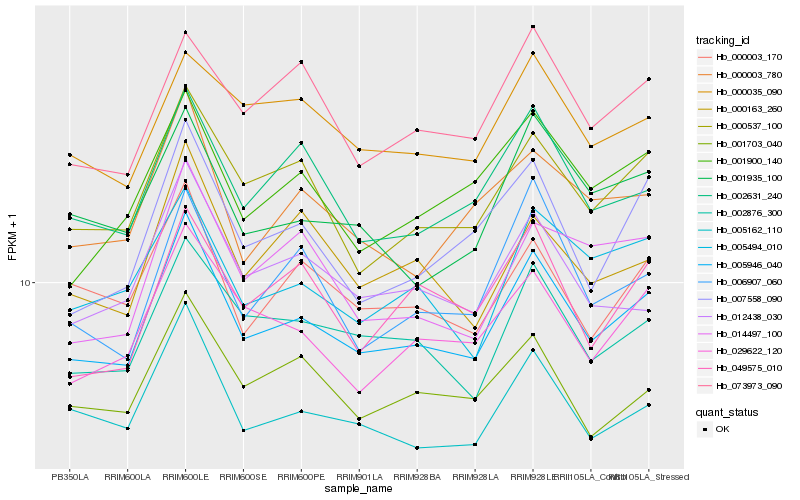
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005946_040 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000537_100 |
0.0642237135 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_005162_110 |
0.0654645205 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 4 |
Hb_002876_300 |
0.0677054666 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_029622_120 |
0.0711505329 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 6 |
Hb_002631_240 |
0.0738617375 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 7 |
Hb_007558_090 |
0.0782412813 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_005494_010 |
0.0789724742 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_001703_040 |
0.0797866394 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000003_170 |
0.0798342271 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 11 |
Hb_001935_100 |
0.0804254427 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 12 |
Hb_012438_030 |
0.0804324401 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 13 |
Hb_001900_140 |
0.0808004159 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000003_780 |
0.0814950261 |
- |
- |
hexokinase [Manihot esculenta] |
| 15 |
Hb_073973_090 |
0.0827737377 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 16 |
Hb_000163_260 |
0.0829637495 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 17 |
Hb_006907_060 |
0.084289607 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000035_090 |
0.0860708885 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 19 |
Hb_014497_100 |
0.087193224 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 20 |
Hb_049575_010 |
0.0881443651 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |