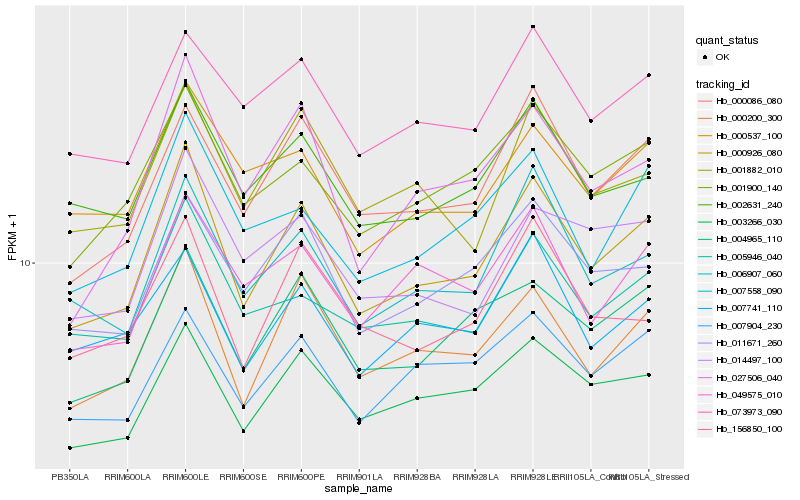
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000926_080 |
0.0 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 2 |
Hb_000200_300 |
0.063141156 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 3 |
Hb_003266_030 |
0.0748541747 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 4 |
Hb_000086_080 |
0.0784359197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004965_110 |
0.0786465191 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 6 |
Hb_006907_060 |
0.0793266723 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_027506_040 |
0.0802219893 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001900_140 |
0.0822490968 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_011671_260 |
0.0835386371 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007558_090 |
0.0860900985 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_002631_240 |
0.0869815604 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 12 |
Hb_005946_040 |
0.0882044853 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001882_010 |
0.0886554278 |
- |
- |
- |
| 14 |
Hb_007904_230 |
0.0908357487 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 15 |
Hb_049575_010 |
0.0909797649 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 16 |
Hb_014497_100 |
0.0916377818 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 17 |
Hb_156850_100 |
0.0928105477 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 18 |
Hb_073973_090 |
0.0945016425 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 19 |
Hb_000537_100 |
0.0947159 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_007741_110 |
0.0952102969 |
- |
- |
- |