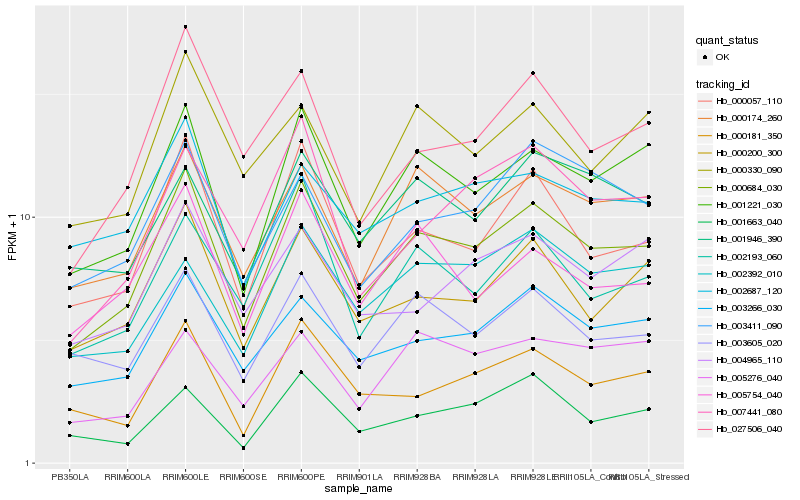
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_030 |
0.0 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 2 |
Hb_001221_030 |
0.0659153447 |
- |
- |
PREDICTED: adenylate kinase 4 [Jatropha curcas] |
| 3 |
Hb_000057_110 |
0.076094111 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 4 |
Hb_000174_260 |
0.0784204517 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent) [Jatropha curcas] |
| 5 |
Hb_003266_030 |
0.0801196972 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 6 |
Hb_000200_300 |
0.0848688452 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 7 |
Hb_003605_020 |
0.0902370991 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 8 |
Hb_027506_040 |
0.0986297067 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005754_040 |
0.0998488096 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002687_120 |
0.1003123532 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000181_350 |
0.100517145 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 12 |
Hb_002193_060 |
0.1018548623 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 13 |
Hb_003411_090 |
0.1022121363 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 4 of pyruvate dehydrogenase complex |
Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000330_090 |
0.1032503708 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001946_390 |
0.1046804157 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 16 |
Hb_007441_080 |
0.1053050607 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 17 |
Hb_004965_110 |
0.1063120736 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 18 |
Hb_002392_010 |
0.1072714794 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 19 |
Hb_005276_040 |
0.1077503176 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 20 |
Hb_001663_040 |
0.1082847942 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |