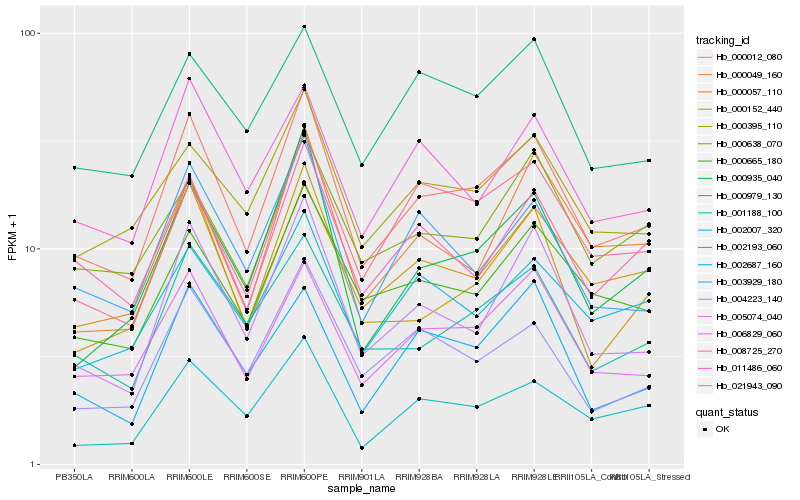
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_080 |
0.0 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 2 |
Hb_006829_060 |
0.073799195 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000057_110 |
0.0864153282 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 4 |
Hb_001188_100 |
0.0884295316 |
- |
- |
hypothetical protein JCGZ_05568 [Jatropha curcas] |
| 5 |
Hb_005074_040 |
0.0885559223 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000665_180 |
0.0935598312 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 7 |
Hb_000395_110 |
0.0939421826 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 8 |
Hb_000935_040 |
0.0947266602 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 9 |
Hb_003929_180 |
0.0960935283 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 10 |
Hb_011486_060 |
0.0963993986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002007_320 |
0.0966871887 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 12 |
Hb_004223_140 |
0.0980996678 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 13 |
Hb_008725_270 |
0.0985309325 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 14 |
Hb_000152_440 |
0.1008188056 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 15 |
Hb_002193_060 |
0.101952451 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 16 |
Hb_000638_070 |
0.1081356943 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 17 |
Hb_000979_130 |
0.1102331962 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 18 |
Hb_021943_090 |
0.1111664145 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 19 |
Hb_000049_160 |
0.1118404488 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 20 |
Hb_002687_160 |
0.1122140105 |
- |
- |
PREDICTED: uncharacterized protein LOC105643395 [Jatropha curcas] |