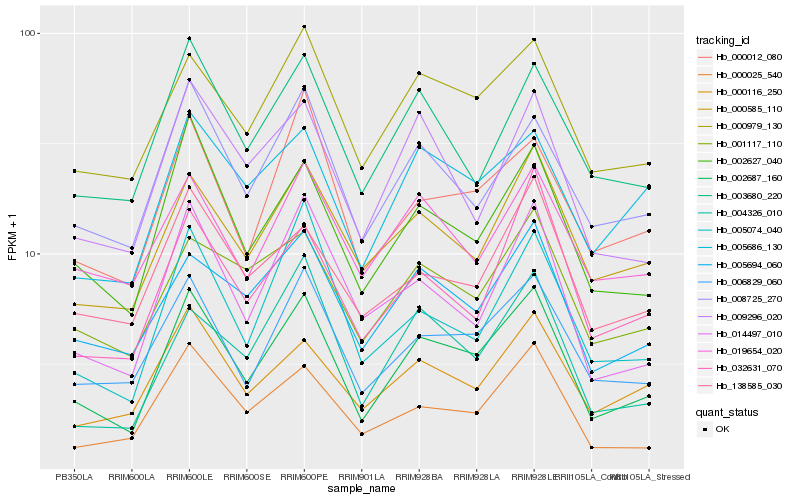
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002687_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643395 [Jatropha curcas] |
| 2 |
Hb_002627_040 |
0.0894133362 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 3 |
Hb_006829_060 |
0.0922688748 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000979_130 |
0.1023821267 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_000025_540 |
0.1040864741 |
- |
- |
PREDICTED: uncharacterized protein LOC104879644 [Vitis vinifera] |
| 6 |
Hb_008725_270 |
0.1058971563 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 7 |
Hb_014497_010 |
0.106451721 |
- |
- |
PREDICTED: uncharacterized protein LOC105635153 [Jatropha curcas] |
| 8 |
Hb_005694_060 |
0.1074696216 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 9 |
Hb_000585_110 |
0.1091037655 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 10 |
Hb_000012_080 |
0.1122140105 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 11 |
Hb_000116_250 |
0.1127955304 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 12 |
Hb_005074_040 |
0.1140790863 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_009296_020 |
0.1149447058 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_003680_220 |
0.1156059789 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 15 |
Hb_001117_110 |
0.1173805879 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 16 |
Hb_032631_070 |
0.117935244 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_005686_130 |
0.1182222467 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 18 |
Hb_004326_010 |
0.1185876883 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_138585_030 |
0.1189789373 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 20 |
Hb_019654_020 |
0.1247377925 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |