| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
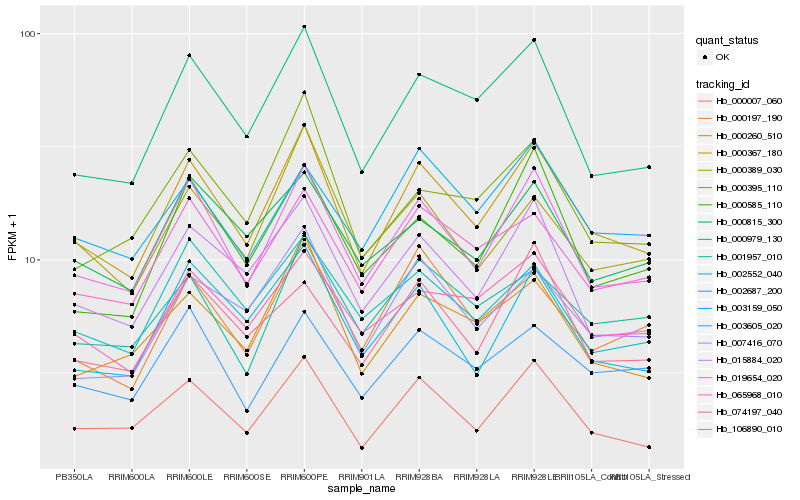
Hb_000367_180 |
0.0 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 2 |
Hb_019654_020 |
0.0525095487 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |
| 3 |
Hb_000007_060 |
0.0641182508 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000979_130 |
0.0675458558 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 5 |
Hb_065968_010 |
0.0758111625 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 6 |
Hb_015884_020 |
0.0760371771 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 7 |
Hb_000260_510 |
0.0780548651 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 8 |
Hb_007416_070 |
0.0821841544 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 9 |
Hb_074197_040 |
0.0834685918 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 10 |
Hb_002687_200 |
0.0884544056 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003605_020 |
0.0888351714 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 12 |
Hb_001957_010 |
0.091056759 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000197_190 |
0.091391641 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000815_300 |
0.0924987131 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 15 |
Hb_002552_040 |
0.0929413906 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 16 |
Hb_003159_050 |
0.0929632657 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_000585_110 |
0.0934409853 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 18 |
Hb_000389_030 |
0.0934989433 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 19 |
Hb_106890_010 |
0.0936664037 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_000395_110 |
0.0943317468 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |