| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
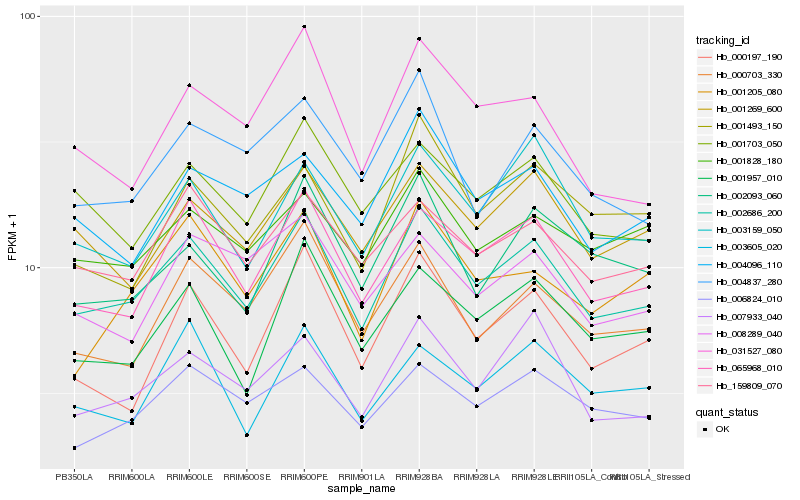
Hb_002686_200 |
0.0 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 2 |
Hb_065968_010 |
0.0605780349 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 3 |
Hb_159809_070 |
0.075710634 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 4 |
Hb_001269_600 |
0.0791833327 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 5 |
Hb_003159_050 |
0.079782602 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 3, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_007933_040 |
0.0798471064 |
- |
- |
PREDICTED: probable beta-D-xylosidase 6 isoform X2 [Jatropha curcas] |
| 7 |
Hb_006824_010 |
0.0803780016 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 8 |
Hb_001703_050 |
0.0805011538 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 9 |
Hb_000703_330 |
0.080628344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001957_010 |
0.0817326873 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004096_110 |
0.084857422 |
- |
- |
PREDICTED: T-complex protein 1 subunit gamma-like [Jatropha curcas] |
| 12 |
Hb_001828_180 |
0.0850733277 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 13 |
Hb_000197_190 |
0.0871759127 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_004837_280 |
0.089243932 |
- |
- |
PREDICTED: uncharacterized protein LOC105648296 [Jatropha curcas] |
| 15 |
Hb_002093_060 |
0.0909271236 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 16 |
Hb_031527_080 |
0.0910065375 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 17 |
Hb_008289_040 |
0.0911829729 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 18 |
Hb_001493_150 |
0.0923261228 |
- |
- |
PREDICTED: alanine aminotransferase 2-like [Jatropha curcas] |
| 19 |
Hb_001205_080 |
0.0931945035 |
- |
- |
Protein FAM96B, putative [Ricinus communis] |
| 20 |
Hb_003605_020 |
0.0961100839 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |