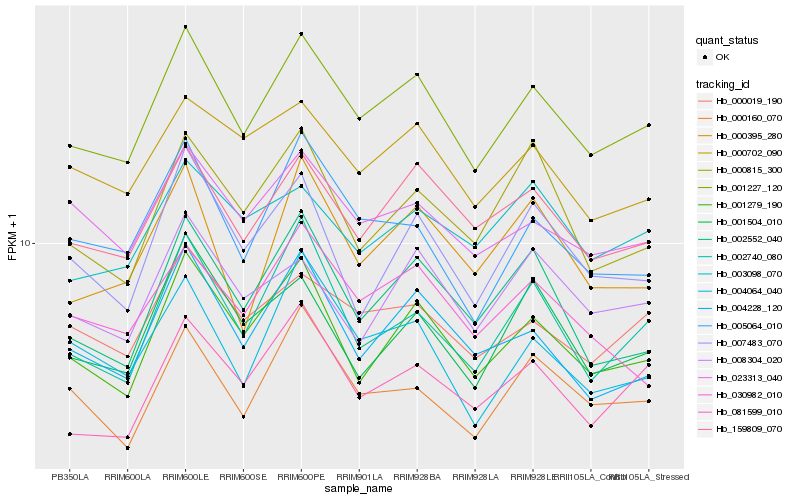
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001227_120 |
0.0 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 2 |
Hb_159809_070 |
0.055379331 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 3 |
Hb_001279_190 |
0.0629378981 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000019_190 |
0.0646355935 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 5 |
Hb_023313_040 |
0.0699633545 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000702_090 |
0.0719562901 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 7 |
Hb_081599_010 |
0.0740673076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004228_120 |
0.0749876573 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 9 |
Hb_005064_010 |
0.0781827794 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 10 |
Hb_008304_020 |
0.0786226701 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 11 |
Hb_002552_040 |
0.0786984568 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 12 |
Hb_003098_070 |
0.07875996 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 13 |
Hb_002740_080 |
0.0788883581 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 14 |
Hb_000160_070 |
0.0792602076 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 15 |
Hb_007483_070 |
0.0816326348 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 16 |
Hb_000815_300 |
0.0817192369 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 17 |
Hb_004064_040 |
0.0825277475 |
- |
- |
PREDICTED: Down syndrome critical region protein 3 homolog isoform X3 [Jatropha curcas] |
| 18 |
Hb_000395_280 |
0.0828812446 |
- |
- |
PREDICTED: thioredoxin-related transmembrane protein 2 [Vitis vinifera] |
| 19 |
Hb_001504_010 |
0.0830678924 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 20 |
Hb_030982_010 |
0.0833828542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |