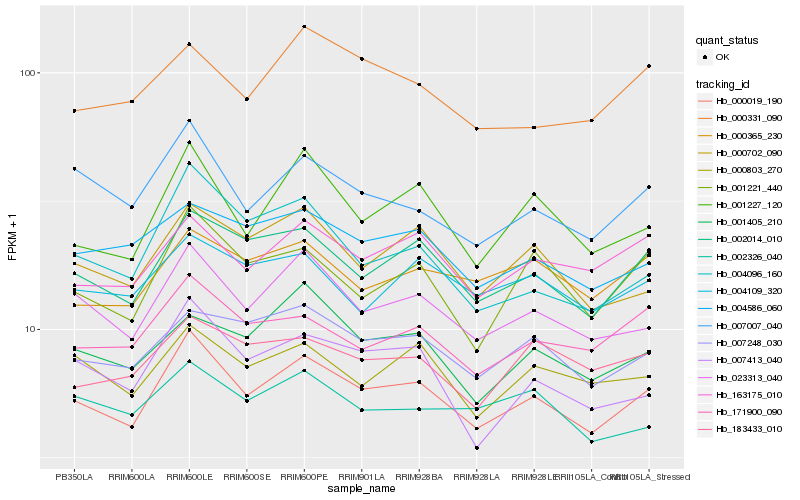
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000019_190 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 2 |
Hb_023313_040 |
0.0516727286 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002014_010 |
0.0529711941 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_001227_120 |
0.0646355935 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 5 |
Hb_007007_040 |
0.065345409 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 6 |
Hb_007248_030 |
0.0666219683 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 7 |
Hb_163175_010 |
0.0678499857 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 8 |
Hb_001221_440 |
0.0687635437 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_171900_090 |
0.0690357182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000702_090 |
0.0690755731 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 11 |
Hb_007413_040 |
0.0722659597 |
- |
- |
myo inositol monophosphatase, putative [Ricinus communis] |
| 12 |
Hb_000803_270 |
0.0723741124 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 13 |
Hb_004586_060 |
0.0732779676 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 14 |
Hb_001405_210 |
0.0748415935 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 15 |
Hb_000365_230 |
0.0772674024 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 16 |
Hb_004109_320 |
0.077562569 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 17 |
Hb_183433_010 |
0.0792692985 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 18 |
Hb_004096_160 |
0.080950349 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 19 |
Hb_000331_090 |
0.0816078631 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 20 |
Hb_002326_040 |
0.0816516451 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |