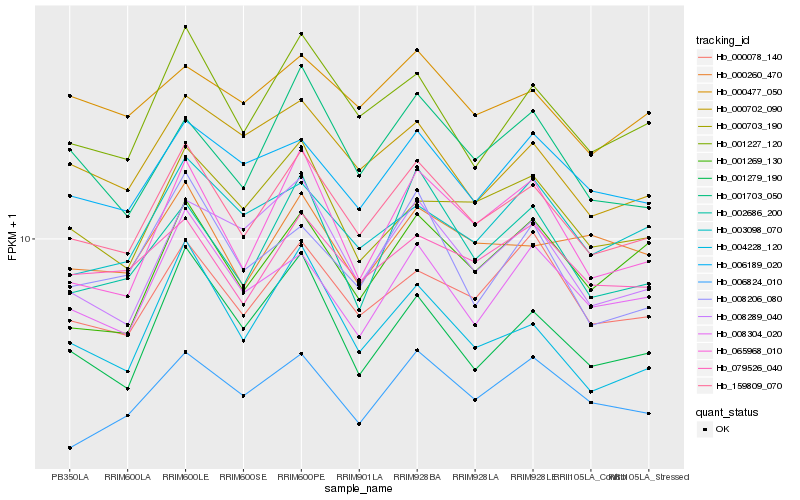
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_159809_070 |
0.0 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 2 |
Hb_001227_120 |
0.055379331 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 3 |
Hb_000078_140 |
0.0623997762 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 4 |
Hb_079526_040 |
0.0651637437 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 5 |
Hb_065968_010 |
0.0653874324 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 6 |
Hb_006189_020 |
0.0665328529 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 7 |
Hb_008304_020 |
0.0687993854 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 8 |
Hb_000702_090 |
0.0704127881 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 9 |
Hb_000703_190 |
0.0704245692 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008289_040 |
0.0716526723 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 11 |
Hb_003098_070 |
0.0725549027 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 12 |
Hb_001703_050 |
0.073673338 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 13 |
Hb_000260_470 |
0.0740465813 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 14 |
Hb_004228_120 |
0.0747837823 |
- |
- |
hypothetical protein POPTR_0013s02080g [Populus trichocarpa] |
| 15 |
Hb_006824_010 |
0.0751035136 |
- |
- |
PREDICTED: origin of replication complex subunit 4 [Jatropha curcas] |
| 16 |
Hb_002686_200 |
0.075710634 |
- |
- |
glucosidase II beta subunit, putative [Ricinus communis] |
| 17 |
Hb_001279_190 |
0.0759111689 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000477_050 |
0.0761865529 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 19 |
Hb_001269_130 |
0.0762349539 |
- |
- |
plant poly(A)+ RNA export protein, putative [Ricinus communis] |
| 20 |
Hb_008206_080 |
0.0777768037 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 17 [Jatropha curcas] |