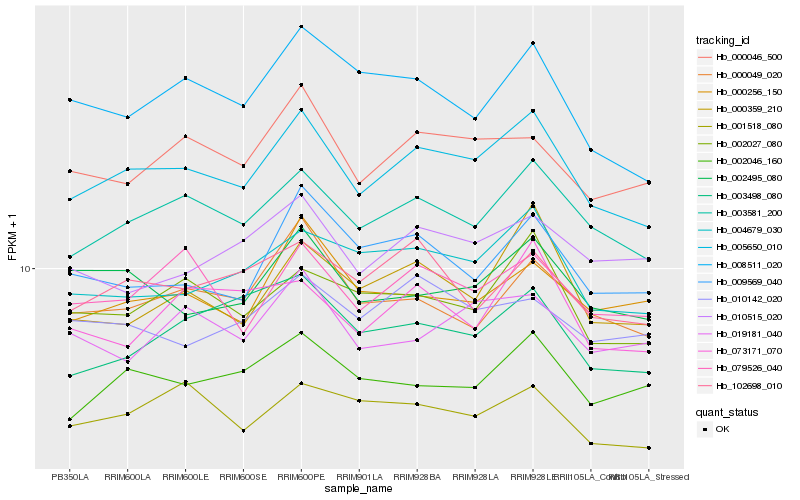
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005650_010 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_003581_200 |
0.0483135535 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 3 |
Hb_004679_030 |
0.0688921164 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 4 |
Hb_002027_080 |
0.0693994419 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 5 |
Hb_000046_500 |
0.0710095435 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 6 |
Hb_079526_040 |
0.0727200231 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 7 |
Hb_000359_210 |
0.0745254397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002495_080 |
0.0748577922 |
- |
- |
PREDICTED: polygalacturonase ADPG2 [Jatropha curcas] |
| 9 |
Hb_000049_020 |
0.0763864627 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 10 |
Hb_102698_010 |
0.0765408532 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002046_160 |
0.0767898319 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_008511_020 |
0.0776694531 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 13 |
Hb_001518_080 |
0.0785209889 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 14 |
Hb_009569_040 |
0.0786465729 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 15 |
Hb_000256_150 |
0.080976808 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 16 |
Hb_003498_080 |
0.0824083413 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_019181_040 |
0.0830695029 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 18 |
Hb_010515_020 |
0.0837956407 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 19 |
Hb_073171_070 |
0.0839612553 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 20 |
Hb_010142_020 |
0.0843989905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |