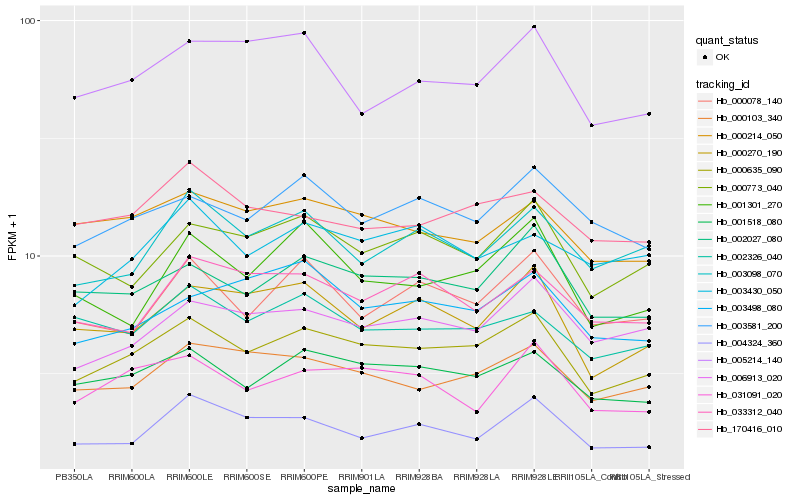
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_090 |
0.0 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 2 |
Hb_001518_080 |
0.0564343024 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 3 |
Hb_002027_080 |
0.0626018091 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 4 |
Hb_000103_340 |
0.0680768955 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 5 |
Hb_006913_020 |
0.0721952942 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000270_190 |
0.0738431494 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 7 |
Hb_000773_040 |
0.0756500603 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003581_200 |
0.0762536274 |
- |
- |
PREDICTED: uncharacterized protein LOC105640179 [Jatropha curcas] |
| 9 |
Hb_000214_050 |
0.0776954213 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_031091_020 |
0.0802649488 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 11 |
Hb_000078_140 |
0.0811858515 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 12 |
Hb_003098_070 |
0.0828214136 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 13 |
Hb_004324_360 |
0.0830292083 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005214_140 |
0.083488793 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 15 |
Hb_002326_040 |
0.0838269757 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 16 |
Hb_003498_080 |
0.0846075396 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 17 |
Hb_170416_010 |
0.0846903275 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 18 |
Hb_001301_270 |
0.0855534088 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 19 |
Hb_003430_050 |
0.0857938303 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 20 |
Hb_033312_040 |
0.0859598553 |
- |
- |
PREDICTED: gamma-tubulin complex component 5-like isoform X1 [Jatropha curcas] |