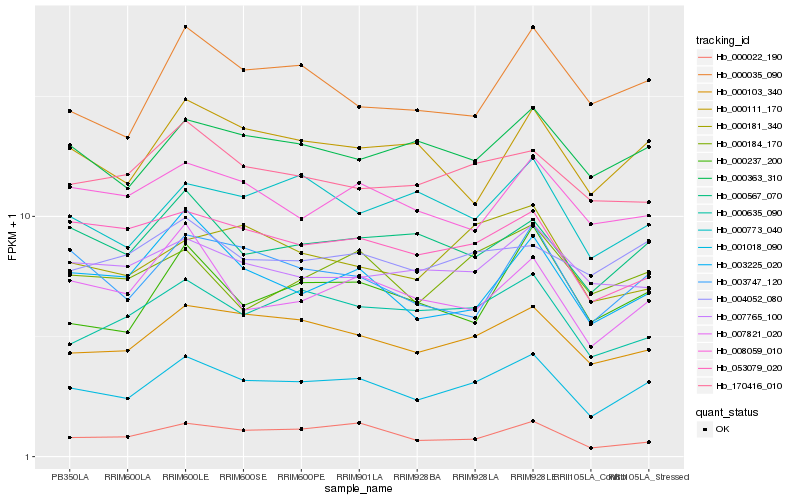
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001018_090 |
0.0 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 2 |
Hb_000103_340 |
0.0683045598 |
- |
- |
PREDICTED: uncharacterized protein LOC105632051 isoform X4 [Jatropha curcas] |
| 3 |
Hb_000363_310 |
0.0839716818 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 4 |
Hb_000567_070 |
0.0855292137 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 5 |
Hb_000635_090 |
0.0871666025 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 6 |
Hb_003747_120 |
0.0875780348 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000111_170 |
0.0908216546 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007821_020 |
0.0912726369 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein DDB_G0275467 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000035_090 |
0.091371927 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 10 |
Hb_003225_020 |
0.0918429674 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 11 |
Hb_170416_010 |
0.0929959873 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 12 |
Hb_000773_040 |
0.0931750022 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004052_080 |
0.094096463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000237_200 |
0.0941952772 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 15 |
Hb_053079_020 |
0.0944874436 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 16 |
Hb_000181_340 |
0.0945985652 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 17 |
Hb_007765_100 |
0.0954245781 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 18 |
Hb_000184_170 |
0.0955890123 |
- |
- |
hypothetical protein RCOM_1213430 [Ricinus communis] |
| 19 |
Hb_008059_010 |
0.095612418 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000022_190 |
0.0957063295 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |