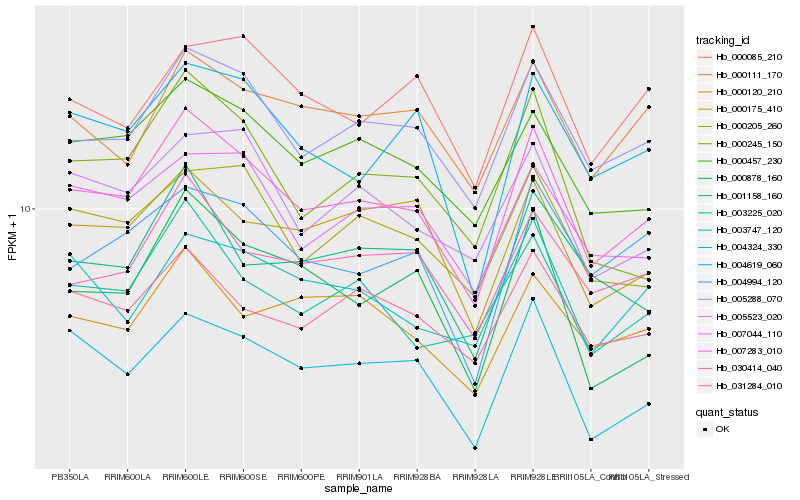
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007283_010 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 2 |
Hb_005288_070 |
0.0499435437 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000457_230 |
0.0618007076 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_030414_040 |
0.0663617338 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004324_330 |
0.0695053365 |
- |
- |
PREDICTED: uncharacterized protein LOC105648374 [Jatropha curcas] |
| 6 |
Hb_031284_010 |
0.0708428477 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 7 |
Hb_000205_260 |
0.070994072 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 8 |
Hb_007044_110 |
0.071963058 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_004619_060 |
0.0737537258 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 10 |
Hb_003225_020 |
0.0739752289 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HAT3.1 [Jatropha curcas] |
| 11 |
Hb_000175_410 |
0.0743789211 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_005523_020 |
0.0746528195 |
- |
- |
MORPHEUS MOLECULE family protein [Populus trichocarpa] |
| 13 |
Hb_004994_120 |
0.0756425198 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 13-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000120_210 |
0.0762292306 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 15 |
Hb_000111_170 |
0.0772577972 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001158_160 |
0.0791689107 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003747_120 |
0.0791740165 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000085_210 |
0.0811353102 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 19 |
Hb_000878_160 |
0.0819313002 |
- |
- |
PREDICTED: D-lactate dehydrogenase [cytochrome], mitochondrial [Jatropha curcas] |
| 20 |
Hb_000245_150 |
0.0823058821 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |