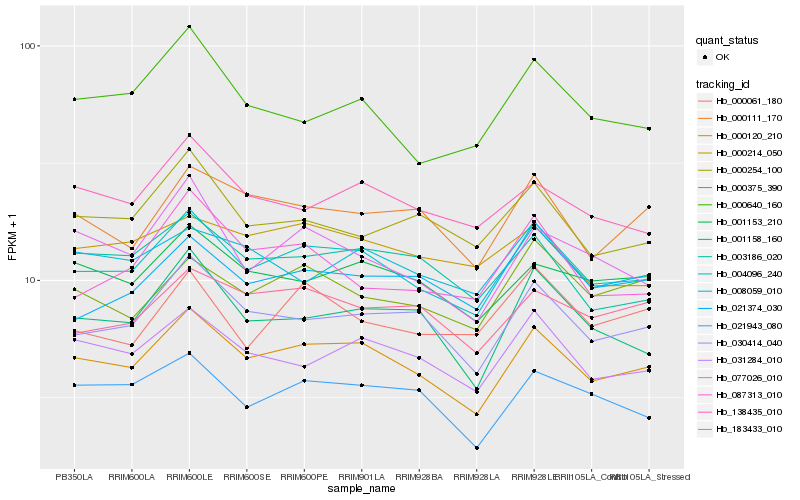
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004096_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 2 |
Hb_000120_210 |
0.0297800802 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 3 |
Hb_030414_040 |
0.0605906498 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_031284_010 |
0.0646719368 |
- |
- |
PREDICTED: uncharacterized protein LOC105633543 [Jatropha curcas] |
| 5 |
Hb_000375_390 |
0.0714036166 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 6 |
Hb_021943_080 |
0.0729303139 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003186_020 |
0.0731926296 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001158_160 |
0.0732402743 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X1 [Jatropha curcas] |
| 9 |
Hb_021374_030 |
0.0743040173 |
- |
- |
hypothetical protein RCOM_0351490 [Ricinus communis] |
| 10 |
Hb_001153_210 |
0.074711732 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 11 |
Hb_138435_010 |
0.0754482819 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 12 |
Hb_000640_160 |
0.0758286491 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 13 |
Hb_183433_010 |
0.0768789311 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 14 |
Hb_000254_100 |
0.076897351 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 15 |
Hb_077026_010 |
0.0780336638 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 16 |
Hb_000111_170 |
0.0786300573 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008059_010 |
0.0788891862 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 18 |
Hb_087313_010 |
0.0799725018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000061_180 |
0.0804093081 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 20 |
Hb_000214_050 |
0.0807200001 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |