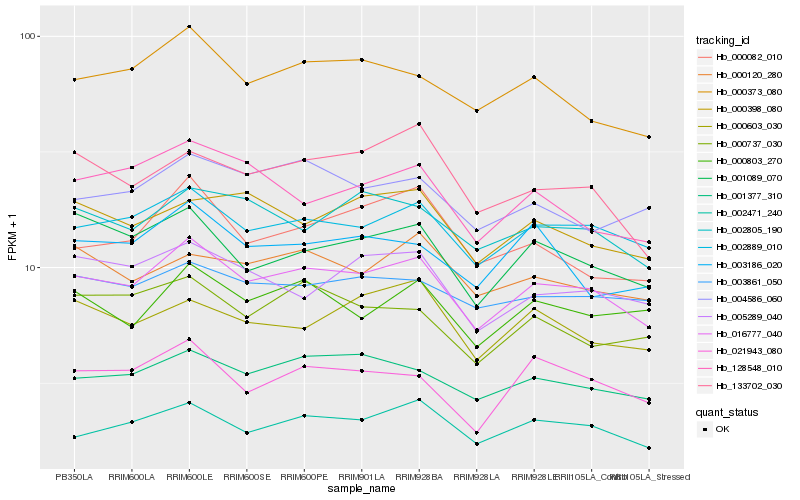
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016777_040 |
0.0 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 2 |
Hb_002889_010 |
0.0471222948 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 3 |
Hb_002471_240 |
0.0579544146 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001377_310 |
0.0584572949 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 5 |
Hb_000373_080 |
0.0634806757 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 6 |
Hb_001089_070 |
0.0667899347 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 7 |
Hb_002805_190 |
0.067961314 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 8 |
Hb_000803_270 |
0.0692734948 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 9 |
Hb_003861_050 |
0.0714776035 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 10 |
Hb_021943_080 |
0.0720389496 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005289_040 |
0.0722489598 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 12 |
Hb_133702_030 |
0.0734380222 |
- |
- |
PREDICTED: uric acid degradation bifunctional protein TTL isoform X1 [Jatropha curcas] |
| 13 |
Hb_000120_280 |
0.0734896473 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 14 |
Hb_004586_060 |
0.0744402812 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 15 |
Hb_000398_080 |
0.0745065643 |
- |
- |
tip120, putative [Ricinus communis] |
| 16 |
Hb_000737_030 |
0.0751208707 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000603_030 |
0.0764160124 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 18 |
Hb_003186_020 |
0.0766792284 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000082_010 |
0.076698608 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 20 |
Hb_128548_010 |
0.0770270137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |