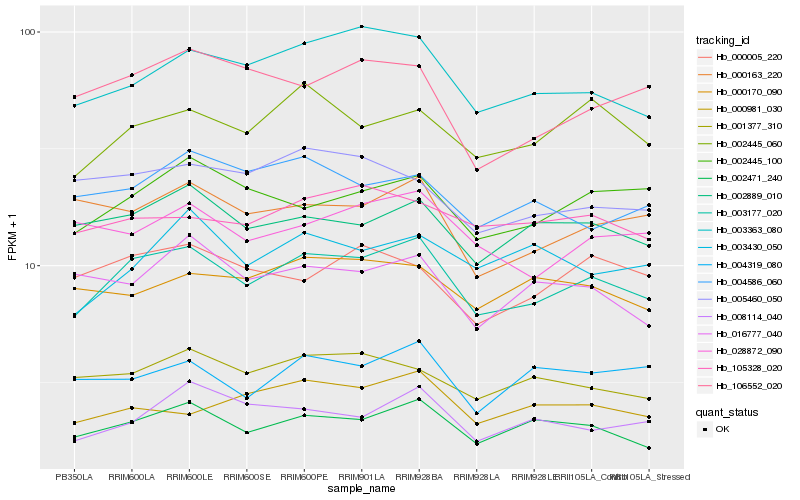
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003177_020 |
0.0 |
- |
- |
diphthine synthase, putative [Ricinus communis] |
| 2 |
Hb_002471_240 |
0.0653216855 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003363_080 |
0.0726140432 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 4 |
Hb_002445_060 |
0.0782110501 |
- |
- |
arf gtpase-activating protein, putative [Ricinus communis] |
| 5 |
Hb_002889_010 |
0.0789322327 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 6 |
Hb_002445_100 |
0.0822168397 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001377_310 |
0.0859468923 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 8 |
Hb_028872_090 |
0.0884127545 |
- |
- |
PREDICTED: urease [Jatropha curcas] |
| 9 |
Hb_008114_040 |
0.0885072706 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 10 |
Hb_105328_020 |
0.0894129658 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 11 |
Hb_000163_220 |
0.0902594628 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 12 |
Hb_000981_030 |
0.0910817411 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 13 |
Hb_016777_040 |
0.0920143263 |
- |
- |
PREDICTED: DNA polymerase eta-like [Jatropha curcas] |
| 14 |
Hb_000170_090 |
0.092663856 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 15 |
Hb_106552_020 |
0.0931140513 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000005_220 |
0.0933988716 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 17 |
Hb_004319_080 |
0.0934502061 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 18 |
Hb_005460_050 |
0.0936721253 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 19 |
Hb_004586_060 |
0.0944059616 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 20 |
Hb_003430_050 |
0.0950990609 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |