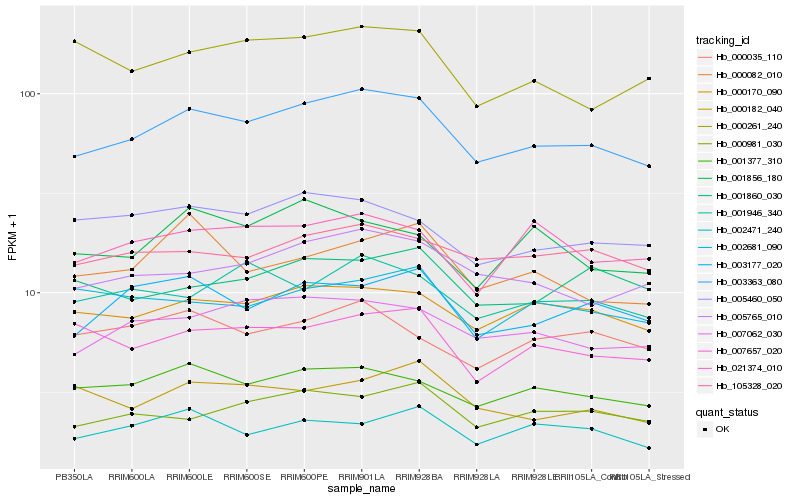
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003363_080 |
0.0 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 2 |
Hb_007062_030 |
0.0648220912 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 3 |
Hb_000170_090 |
0.067493103 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 4 |
Hb_005765_010 |
0.0678421647 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001377_310 |
0.0681345588 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 6 |
Hb_003177_020 |
0.0726140432 |
- |
- |
diphthine synthase, putative [Ricinus communis] |
| 7 |
Hb_000981_030 |
0.0770681216 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 8 |
Hb_105328_020 |
0.0772854732 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 9 |
Hb_005460_050 |
0.0797700202 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 10 |
Hb_007657_020 |
0.0808093489 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 11 |
Hb_001946_340 |
0.0808238644 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_021374_010 |
0.0832718933 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 13 |
Hb_002471_240 |
0.0845428676 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000035_110 |
0.0860972943 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 15 |
Hb_000082_010 |
0.0874144273 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 16 |
Hb_002681_090 |
0.0877378881 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000261_240 |
0.0885579216 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 18 |
Hb_000182_040 |
0.088587957 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 19 |
Hb_001856_180 |
0.0888669701 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 20 |
Hb_001860_030 |
0.090145832 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |