| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
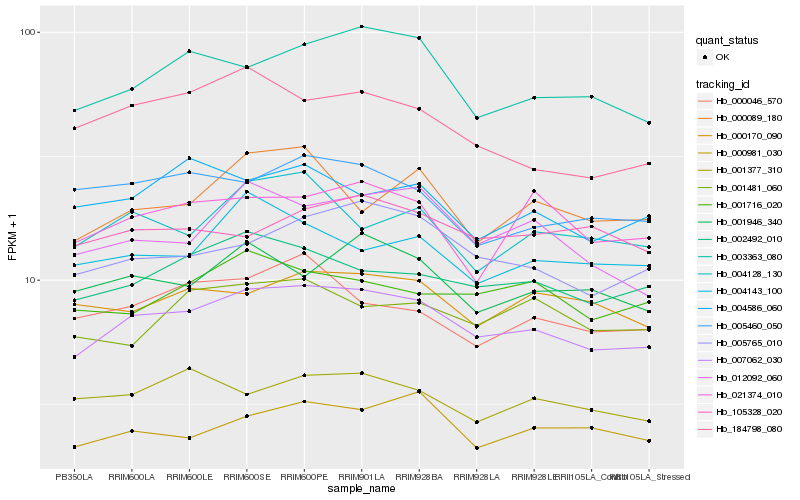
Hb_007062_030 |
0.0 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 2 |
Hb_005765_010 |
0.0631187001 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002492_010 |
0.0635822955 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 4 |
Hb_003363_080 |
0.0648220912 |
- |
- |
eukaryotic translation elongation factor 1B gamma-subunit [Hevea brasiliensis] |
| 5 |
Hb_001946_340 |
0.0721668349 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_012092_060 |
0.0729132505 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 7 |
Hb_000170_090 |
0.0748882993 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 8 |
Hb_000981_030 |
0.0753192832 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 9 |
Hb_001716_020 |
0.0764931662 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 10 |
Hb_000046_570 |
0.0767817593 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 11 |
Hb_004128_130 |
0.0770895278 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001481_060 |
0.0775155583 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_184798_080 |
0.0777372486 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 14 |
Hb_021374_010 |
0.0783724763 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 15 |
Hb_001377_310 |
0.0786485861 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 16 |
Hb_004586_060 |
0.0790378898 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 17 |
Hb_004143_100 |
0.0791531029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000089_180 |
0.0792803261 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 19 |
Hb_005460_050 |
0.0803763595 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 20 |
Hb_105328_020 |
0.080545236 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |