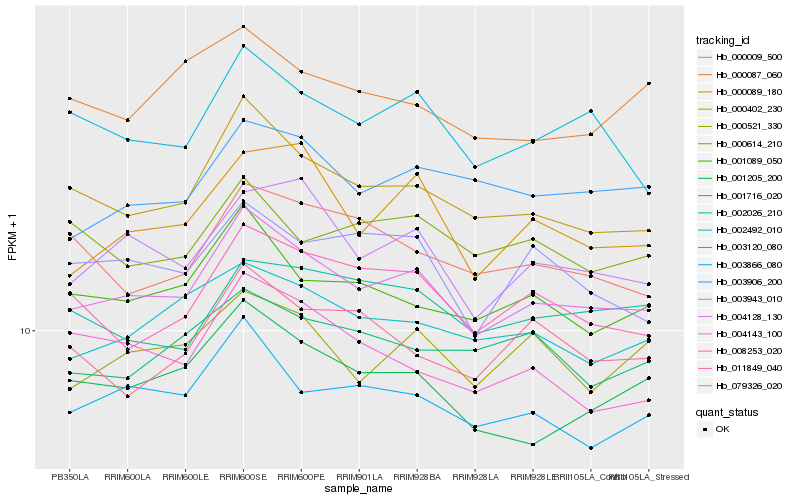
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004143_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000402_230 |
0.0427077122 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_003866_080 |
0.0535969659 |
- |
- |
PREDICTED: uncharacterized protein LOC105639150 [Jatropha curcas] |
| 4 |
Hb_004128_130 |
0.054351811 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001089_050 |
0.0547095922 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 6 |
Hb_003120_080 |
0.0569763214 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002492_010 |
0.057011576 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 8 |
Hb_001205_200 |
0.0592339603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011849_040 |
0.0597665331 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_000089_180 |
0.0605972096 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 11 |
Hb_003906_200 |
0.0610380396 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 12 |
Hb_000521_330 |
0.0623752149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003943_010 |
0.0648769658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002026_210 |
0.0678013578 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 15 |
Hb_000009_500 |
0.0681467162 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 16 |
Hb_001716_020 |
0.0683975788 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 17 |
Hb_000614_210 |
0.0693752569 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000087_060 |
0.0697663177 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 19 |
Hb_079326_020 |
0.0707325756 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008253_020 |
0.0709131556 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |