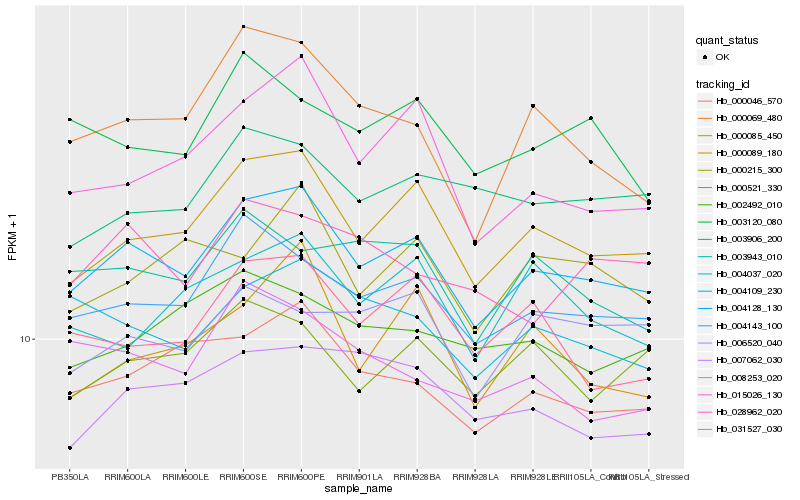
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004128_130 |
0.0 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000089_180 |
0.0434889389 |
- |
- |
PREDICTED: homoserine kinase-like [Jatropha curcas] |
| 3 |
Hb_004143_100 |
0.054351811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000046_570 |
0.0651837999 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 5 |
Hb_015026_130 |
0.0706632901 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 6 |
Hb_004109_230 |
0.0726550455 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_000521_330 |
0.0740746754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000069_480 |
0.075372209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_031527_030 |
0.0756982971 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 10 |
Hb_007062_030 |
0.0770895278 |
- |
- |
hypothetical protein VITISV_039033 [Vitis vinifera] |
| 11 |
Hb_003943_010 |
0.0788489074 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003906_200 |
0.0796448531 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 13 |
Hb_000215_300 |
0.0797685847 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 14 |
Hb_000085_450 |
0.0799626154 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 15 |
Hb_002492_010 |
0.0801498084 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 16 |
Hb_003120_080 |
0.0812377413 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_028962_020 |
0.0820861224 |
- |
- |
hypothetical protein POPTR_0016s00660g [Populus trichocarpa] |
| 18 |
Hb_004037_020 |
0.0823078569 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006520_040 |
0.0829518903 |
- |
- |
PREDICTED: uncharacterized protein LOC105641714 isoform X2 [Jatropha curcas] |
| 20 |
Hb_008253_020 |
0.0831739883 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |