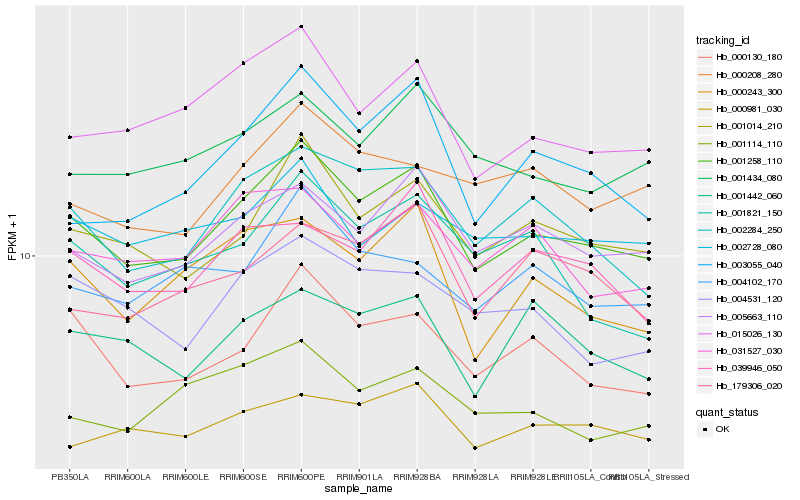
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001258_110 |
0.0 |
- |
- |
transporter, putative [Ricinus communis] |
| 2 |
Hb_001014_210 |
0.060769144 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 3 |
Hb_002284_250 |
0.0685798159 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 4 |
Hb_005663_110 |
0.0705819158 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000130_180 |
0.0727660518 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_015026_130 |
0.074975317 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 7 |
Hb_001434_080 |
0.0829708098 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 8 |
Hb_000243_300 |
0.0830734261 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_003055_040 |
0.0835416768 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_039946_050 |
0.0838421809 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_004531_120 |
0.0858946591 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 12 |
Hb_031527_030 |
0.0866535464 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 13 |
Hb_004102_170 |
0.0866684887 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 14 |
Hb_000981_030 |
0.0873746365 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 15 |
Hb_001442_060 |
0.088927223 |
- |
- |
PREDICTED: uncharacterized protein LOC105638091 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000208_280 |
0.0891677583 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 17 |
Hb_179306_020 |
0.0892276848 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002728_080 |
0.0892566742 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 19 |
Hb_001114_110 |
0.0900075127 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 20 |
Hb_001821_150 |
0.0904856974 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |