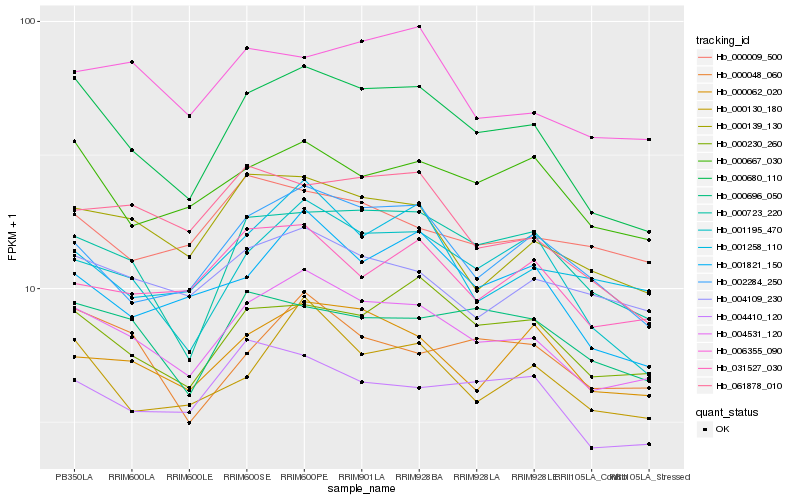
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004531_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 2 |
Hb_000680_110 |
0.0600622661 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 3 |
Hb_000139_130 |
0.0745671635 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 4 |
Hb_004109_230 |
0.0779304685 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_000062_020 |
0.0784255892 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_002284_250 |
0.0803799123 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 7 |
Hb_000130_180 |
0.0812666315 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000230_260 |
0.0822909162 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 9 |
Hb_000723_220 |
0.0826483919 |
- |
- |
PREDICTED: uncharacterized protein LOC105642234 [Jatropha curcas] |
| 10 |
Hb_031527_030 |
0.0841193326 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 11 |
Hb_001821_150 |
0.0845127987 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 12 |
Hb_001195_470 |
0.085201635 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 13 |
Hb_001258_110 |
0.0858946591 |
- |
- |
transporter, putative [Ricinus communis] |
| 14 |
Hb_006355_090 |
0.087142009 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 15 |
Hb_000667_030 |
0.0871645619 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 16 |
Hb_004410_120 |
0.0876234691 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000048_060 |
0.0894414259 |
- |
- |
PREDICTED: probable apyrase 7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000696_050 |
0.0922029897 |
- |
- |
PREDICTED: CSC1-like protein At4g35870 [Jatropha curcas] |
| 19 |
Hb_061878_010 |
0.0927320963 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 20 |
Hb_000009_500 |
0.0928932461 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |