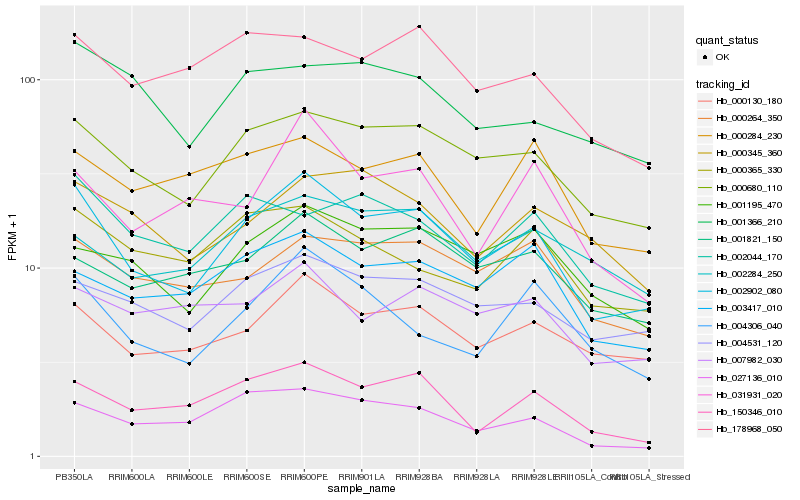
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002902_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 2 |
Hb_000680_110 |
0.0862355961 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 3 |
Hb_000130_180 |
0.103250782 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_004531_120 |
0.1179388349 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 5 |
Hb_150346_010 |
0.1187983742 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_004306_040 |
0.1201972721 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 7 |
Hb_027136_010 |
0.1212651672 |
- |
- |
hypothetical protein L484_010426 [Morus notabilis] |
| 8 |
Hb_001195_470 |
0.123296915 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 9 |
Hb_003417_010 |
0.1256191484 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 10 |
Hb_000264_350 |
0.1261766538 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 11 |
Hb_001821_150 |
0.1276923903 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 12 |
Hb_178968_050 |
0.1327209603 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 13 |
Hb_031931_020 |
0.1339410195 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 14 |
Hb_000365_330 |
0.1347721898 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_002284_250 |
0.1348109474 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 16 |
Hb_007982_030 |
0.1358262073 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g10020 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000284_230 |
0.1363088311 |
- |
- |
PREDICTED: uncharacterized protein At5g49945-like [Jatropha curcas] |
| 18 |
Hb_002044_170 |
0.1368076671 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 19 |
Hb_000345_360 |
0.1369654841 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 20 |
Hb_001366_210 |
0.1377660697 |
- |
- |
cathepsin B, putative [Ricinus communis] |