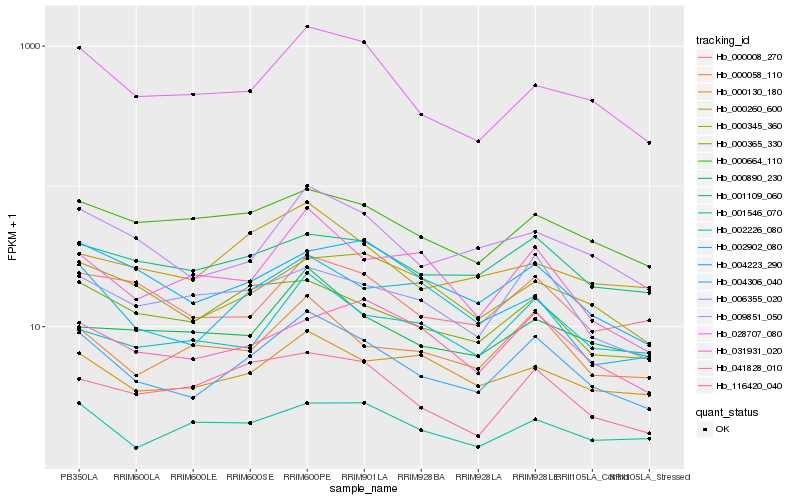
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004306_040 |
0.0 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 2 |
Hb_009851_050 |
0.1101154584 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 3 |
Hb_028707_080 |
0.1156717555 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 4 |
Hb_002902_080 |
0.1201972721 |
- |
- |
PREDICTED: uncharacterized protein LOC105639821 [Jatropha curcas] |
| 5 |
Hb_000058_110 |
0.1216343931 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000365_330 |
0.1279959559 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_000008_270 |
0.129505631 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein REVOLUTA [Jatropha curcas] |
| 8 |
Hb_000130_180 |
0.1312075879 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_031931_020 |
0.1317028858 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 10 |
Hb_000345_360 |
0.1327084343 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 11 |
Hb_004223_290 |
0.1334593307 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 12 |
Hb_006355_020 |
0.1347187953 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 13 |
Hb_002226_080 |
0.136028818 |
- |
- |
PREDICTED: uncharacterized protein LOC105641402 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001109_060 |
0.1370461522 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000260_600 |
0.1400707241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000664_110 |
0.14066245 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 17 |
Hb_000890_230 |
0.1407228481 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 18 |
Hb_041828_010 |
0.1427820033 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 19 |
Hb_116420_040 |
0.143455741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001546_070 |
0.1437829242 |
- |
- |
Uncharacterized protein F383_19872 [Gossypium arboreum] |