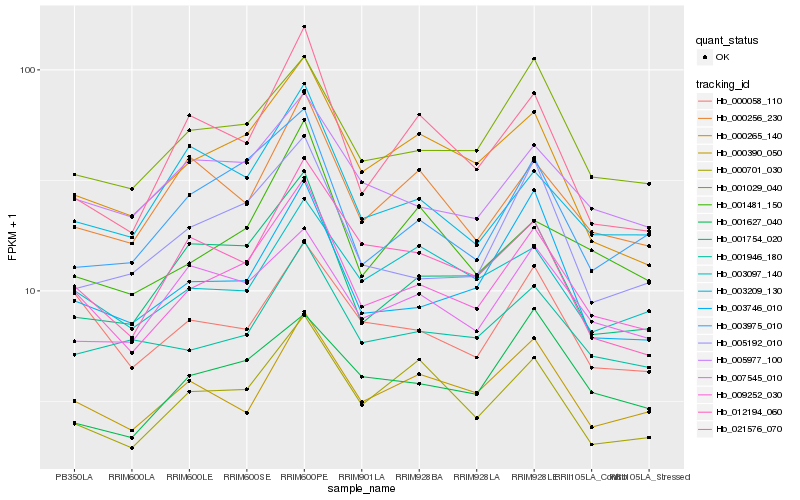
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009252_030 |
0.0 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 2 |
Hb_001029_040 |
0.087916884 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 3 |
Hb_001754_020 |
0.0897475406 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 4 |
Hb_005977_100 |
0.0912600985 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 5 |
Hb_000265_140 |
0.092237566 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |
| 6 |
Hb_001946_180 |
0.0937451876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000058_110 |
0.0980537102 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000390_050 |
0.0981176128 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 9 |
Hb_003746_010 |
0.0984546283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000701_030 |
0.0988314515 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 11 |
Hb_003975_010 |
0.0996030452 |
- |
- |
PREDICTED: protein RTF2 homolog [Jatropha curcas] |
| 12 |
Hb_005192_010 |
0.1010069328 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_001481_150 |
0.102308167 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 14 |
Hb_000256_230 |
0.1040696984 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 15 |
Hb_021576_070 |
0.1072986831 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 16 |
Hb_007545_010 |
0.1080300012 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 17 |
Hb_012194_060 |
0.1083781448 |
- |
- |
PREDICTED: uncharacterized protein LOC105636108 [Jatropha curcas] |
| 18 |
Hb_003209_130 |
0.1087084571 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 19 |
Hb_001627_040 |
0.1104739074 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003097_140 |
0.1120698114 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |