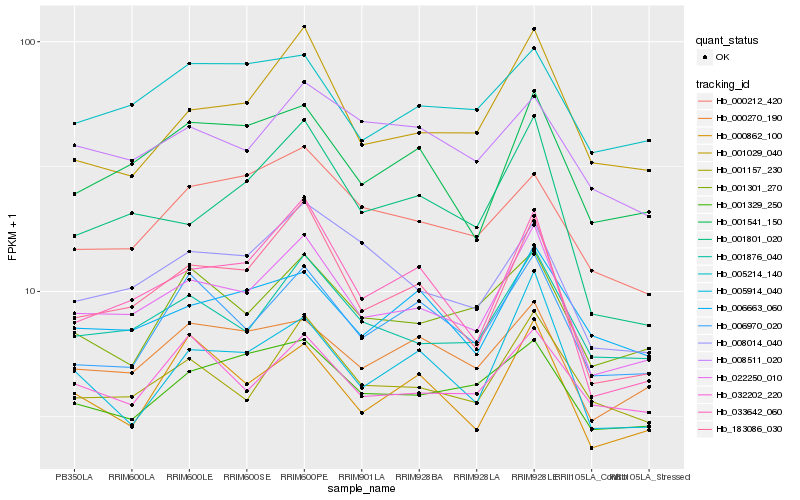
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022250_010 |
0.0 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 2 |
Hb_001876_040 |
0.0587052437 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 3 |
Hb_183086_030 |
0.0657861231 |
- |
- |
GTPase-activating protein GYP7 [Gossypium arboreum] |
| 4 |
Hb_001157_230 |
0.0693332945 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 5 |
Hb_001541_150 |
0.0779160462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001029_040 |
0.0793681435 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 7 |
Hb_000270_190 |
0.0795412608 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 8 |
Hb_033642_060 |
0.0810685216 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_008014_040 |
0.0813758371 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 10 |
Hb_001329_250 |
0.0841848 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006970_020 |
0.0851866671 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 12 |
Hb_001801_020 |
0.0862367267 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_001301_270 |
0.0865247305 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 14 |
Hb_000862_100 |
0.0866828783 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_006663_060 |
0.088085128 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 16 |
Hb_032202_220 |
0.0886642631 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_005914_040 |
0.0892348568 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 18 |
Hb_005214_140 |
0.0904697717 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 19 |
Hb_000212_420 |
0.0913147327 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 20 |
Hb_008511_020 |
0.091523741 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |