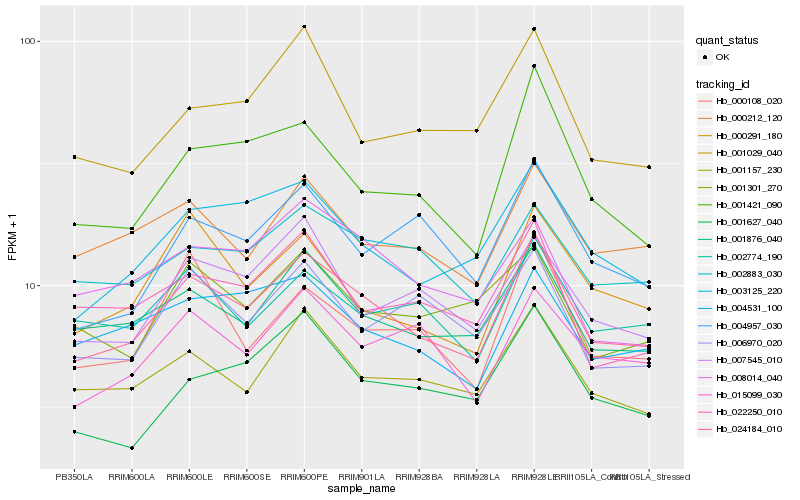
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024184_010 |
0.0 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 2 |
Hb_001876_040 |
0.0704474746 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 3 |
Hb_001157_230 |
0.075690462 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 4 |
Hb_003125_220 |
0.0790558948 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 5 |
Hb_000212_120 |
0.0840391685 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 6 |
Hb_002883_030 |
0.0853983136 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 7 |
Hb_007545_010 |
0.087005066 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 8 |
Hb_015099_030 |
0.0872885487 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 9 |
Hb_004531_100 |
0.0877831798 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002774_190 |
0.0879389898 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 11 |
Hb_001421_090 |
0.0895973568 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 12 |
Hb_022250_010 |
0.0933498949 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 13 |
Hb_006970_020 |
0.0945328767 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 14 |
Hb_000291_180 |
0.0949146717 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 15 |
Hb_008014_040 |
0.0958268718 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 16 |
Hb_001029_040 |
0.0964198446 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 17 |
Hb_004957_030 |
0.0964493277 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 18 |
Hb_000108_020 |
0.0968231064 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 19 |
Hb_001301_270 |
0.0969567437 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 20 |
Hb_001627_040 |
0.0974505705 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 isoform X1 [Jatropha curcas] |