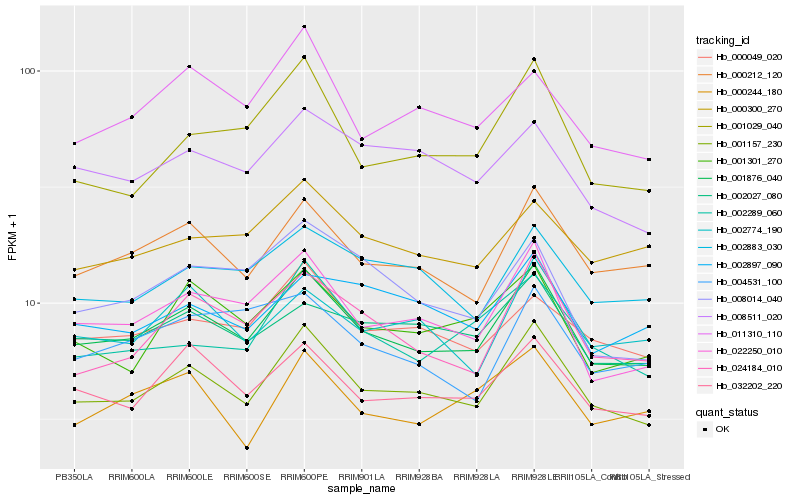
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001876_040 |
0.0 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 2 |
Hb_001157_230 |
0.0354596037 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 3 |
Hb_022250_010 |
0.0587052437 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 4 |
Hb_000212_120 |
0.0590767386 |
- |
- |
hypothetical protein JCGZ_18070 [Jatropha curcas] |
| 5 |
Hb_032202_220 |
0.0647016916 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 6 |
Hb_024184_010 |
0.0704474746 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 7 |
Hb_002027_080 |
0.0756336265 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 8 |
Hb_001301_270 |
0.076989715 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 9 |
Hb_000049_020 |
0.0782896384 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 10 |
Hb_002883_030 |
0.0798172588 |
- |
- |
PREDICTED: uncharacterized protein LOC105638222 [Jatropha curcas] |
| 11 |
Hb_000300_270 |
0.0805480879 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_002774_190 |
0.0805934366 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 13 |
Hb_002289_060 |
0.0817085527 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008511_020 |
0.083981426 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 15 |
Hb_001029_040 |
0.0840755586 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 16 |
Hb_002897_090 |
0.085108023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011310_110 |
0.0864296368 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008014_040 |
0.0872656932 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 19 |
Hb_004531_100 |
0.0883297217 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000244_180 |
0.0899463628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |