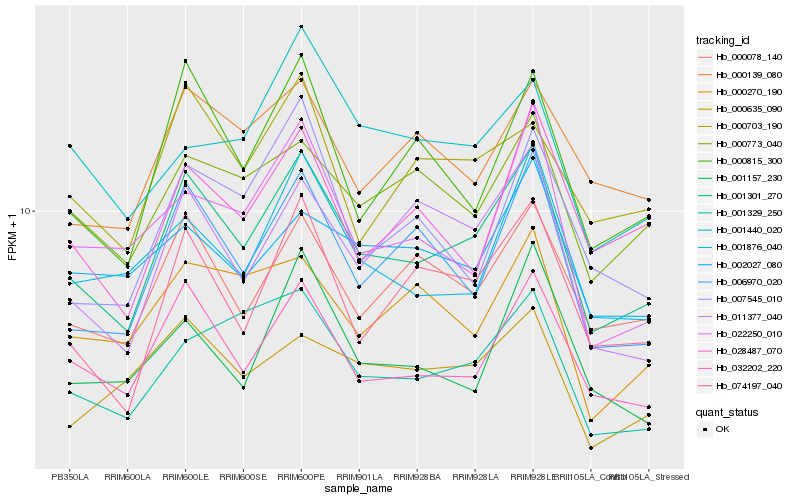
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001301_270 |
0.0 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 2 |
Hb_032202_220 |
0.0584571604 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 3 |
Hb_006970_020 |
0.0677749147 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 4 |
Hb_074197_040 |
0.0691061522 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 5 |
Hb_000078_140 |
0.0691106643 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 6 |
Hb_000815_300 |
0.0718799173 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 7 |
Hb_001329_250 |
0.073707367 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000773_040 |
0.0762806068 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 9 |
Hb_011377_040 |
0.0768303315 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 10 |
Hb_001876_040 |
0.076989715 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 11 |
Hb_000703_190 |
0.0796936295 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000139_080 |
0.0817812735 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000270_190 |
0.0843043521 |
transcription factor |
TF Family: DDT |
tRNA ligase, putative [Ricinus communis] |
| 14 |
Hb_028487_070 |
0.0847249597 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001157_230 |
0.0848829849 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 16 |
Hb_000635_090 |
0.0855534088 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 17 |
Hb_022250_010 |
0.0865247305 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 18 |
Hb_002027_080 |
0.0866113599 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 19 |
Hb_007545_010 |
0.0876213571 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 20 |
Hb_001440_020 |
0.0897145656 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |