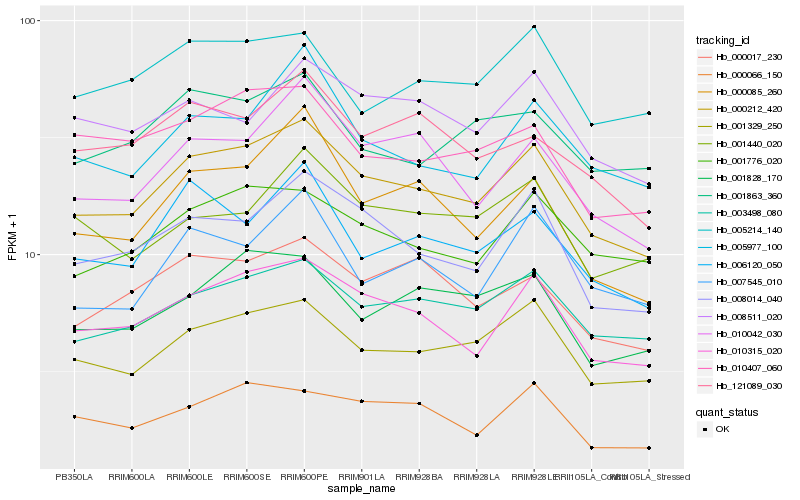
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_420 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 2 |
Hb_010315_020 |
0.0500978785 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 3 |
Hb_001329_250 |
0.0562768595 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003498_080 |
0.0598998846 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 5 |
Hb_008014_040 |
0.0610360743 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 6 |
Hb_001776_020 |
0.0722537425 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 7 |
Hb_010042_030 |
0.0764266559 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine diphosphorylase 1 [Jatropha curcas] |
| 8 |
Hb_000017_230 |
0.0781097178 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 9 |
Hb_005977_100 |
0.0788902782 |
- |
- |
PREDICTED: tetraspanin-18 [Jatropha curcas] |
| 10 |
Hb_006120_050 |
0.0792506098 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 11 |
Hb_001828_170 |
0.0807882024 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 12 |
Hb_000066_150 |
0.0810945062 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 13 |
Hb_007545_010 |
0.0833834955 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 14 |
Hb_000085_260 |
0.0839401138 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 15 |
Hb_121089_030 |
0.084292899 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 16 |
Hb_008511_020 |
0.084422866 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 17 |
Hb_010407_060 |
0.0857925002 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 18 |
Hb_001440_020 |
0.0861613166 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 19 |
Hb_001863_360 |
0.0862824135 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 20 |
Hb_005214_140 |
0.0864082593 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |