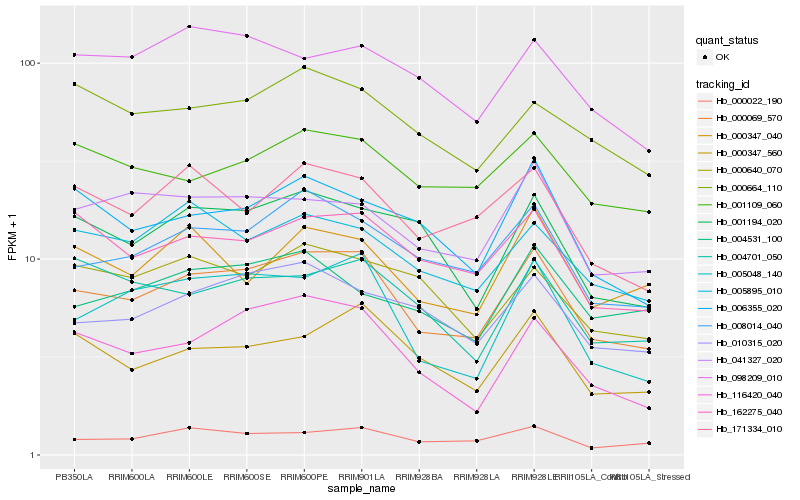
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_570 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_008014_040 |
0.0804003621 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 3 |
Hb_000022_190 |
0.0892606617 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |
| 4 |
Hb_005048_140 |
0.0896205807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_116420_040 |
0.091582816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010315_020 |
0.0924046197 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 7 |
Hb_162275_040 |
0.0924087577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_041327_020 |
0.0961070994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000347_560 |
0.0962699287 |
- |
- |
- |
| 10 |
Hb_001194_020 |
0.0986840458 |
- |
- |
microtubule associated protein xmap215, putative [Ricinus communis] |
| 11 |
Hb_171334_010 |
0.099005478 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 12 |
Hb_005895_010 |
0.1007539178 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006355_020 |
0.1008964361 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 14 |
Hb_001109_060 |
0.1053772609 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000664_110 |
0.1073746179 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 16 |
Hb_004701_050 |
0.1085089749 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 17 |
Hb_000347_040 |
0.1092884067 |
- |
- |
hypothetical protein JCGZ_24660 [Jatropha curcas] |
| 18 |
Hb_000640_070 |
0.110898574 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 19 |
Hb_004531_100 |
0.1114571366 |
- |
- |
PREDICTED: zinc phosphodiesterase ELAC protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_098209_010 |
0.1116734811 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |