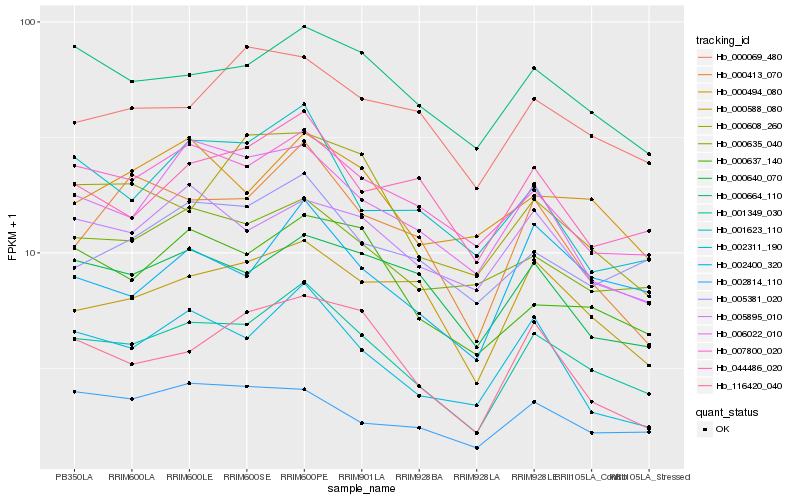
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001349_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000664_110 |
0.0978205391 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 3 |
Hb_007800_020 |
0.098682793 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000637_140 |
0.1019591084 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 5 |
Hb_002400_320 |
0.1031255497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002311_190 |
0.1050895435 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 7 |
Hb_000588_080 |
0.1065974325 |
- |
- |
PREDICTED: uncharacterized protein LOC105638459 [Jatropha curcas] |
| 8 |
Hb_116420_040 |
0.109206443 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000635_040 |
0.1092744227 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 10 |
Hb_044486_020 |
0.1100735603 |
- |
- |
CASTOR protein [Glycine max] |
| 11 |
Hb_005381_020 |
0.111247585 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 12 |
Hb_001623_110 |
0.111391519 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_006022_010 |
0.1128437377 |
- |
- |
PREDICTED: uncharacterized protein LOC105628883 [Jatropha curcas] |
| 14 |
Hb_000640_070 |
0.1130731694 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 15 |
Hb_005895_010 |
0.1131235225 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000413_070 |
0.1133714698 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002814_110 |
0.1137599153 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 18 |
Hb_000494_080 |
0.1161666466 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000608_260 |
0.1171844461 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 20 |
Hb_000069_480 |
0.1180599378 |
- |
- |
conserved hypothetical protein [Ricinus communis] |