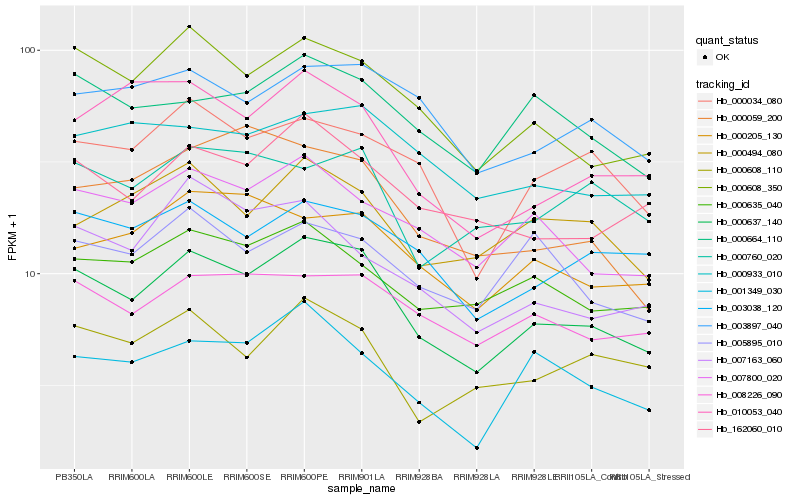
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000637_140 |
0.0 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 2 |
Hb_000608_350 |
0.0766587661 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 3 |
Hb_007800_020 |
0.0924602255 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_162060_010 |
0.0954487303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008226_090 |
0.0989999121 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000034_080 |
0.0998837452 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003038_120 |
0.1006090794 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 8 |
Hb_000635_040 |
0.1011045713 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 9 |
Hb_001349_030 |
0.1019591084 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000664_110 |
0.102118352 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 11 |
Hb_003897_040 |
0.102953189 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 12 |
Hb_000608_110 |
0.1055530089 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 13 |
Hb_000933_010 |
0.1063280425 |
- |
- |
- |
| 14 |
Hb_000205_130 |
0.1067585208 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_010053_040 |
0.1070809688 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 16 |
Hb_000494_080 |
0.1071435044 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_005895_010 |
0.1116447516 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000059_200 |
0.1126302954 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 19 |
Hb_007163_060 |
0.1126834448 |
- |
- |
PREDICTED: protein RMD5 homolog A [Jatropha curcas] |
| 20 |
Hb_000760_020 |
0.1130292894 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |