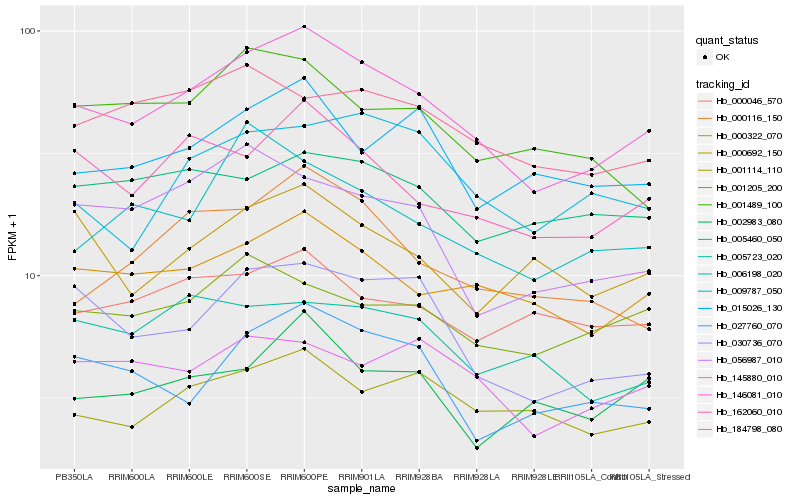
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_146081_010 |
0.0 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 2 |
Hb_000322_070 |
0.0829105946 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_162060_010 |
0.08637873 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_030736_070 |
0.0964282589 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001114_110 |
0.0989662979 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 6 |
Hb_000116_150 |
0.1035077129 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 7 |
Hb_145880_010 |
0.1062608469 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 8 |
Hb_015026_130 |
0.1064871299 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 9 |
Hb_184798_080 |
0.1070720889 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 10 |
Hb_027760_070 |
0.107161748 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 11 |
Hb_001205_200 |
0.1073911887 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009787_050 |
0.1095919745 |
- |
- |
(Di)nucleoside polyphosphate hydrolase, putative [Ricinus communis] |
| 13 |
Hb_006198_020 |
0.1102328949 |
- |
- |
PREDICTED: uncharacterized protein LOC105644416 [Jatropha curcas] |
| 14 |
Hb_000692_150 |
0.1104901346 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 3 [Jatropha curcas] |
| 15 |
Hb_000046_570 |
0.1108898512 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 16 |
Hb_056987_010 |
0.1108899322 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 17 |
Hb_001489_100 |
0.1112875674 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 18 |
Hb_005460_050 |
0.1121878221 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 19 |
Hb_002983_080 |
0.1123549959 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |
| 20 |
Hb_005723_020 |
0.113441351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |