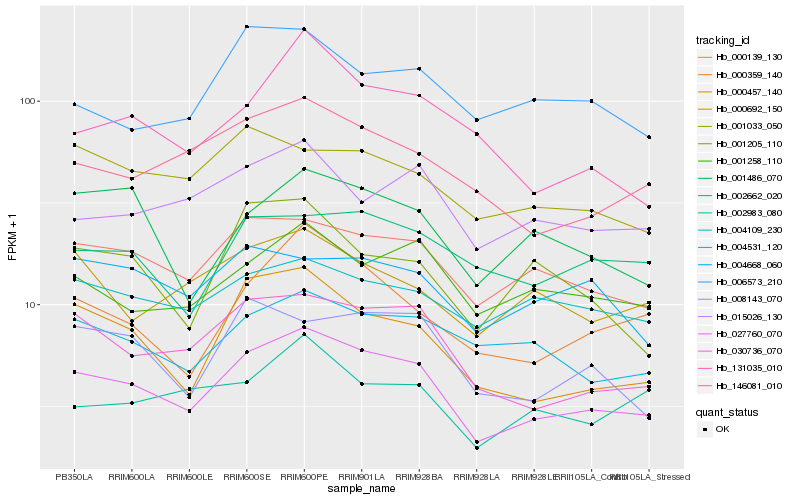
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027760_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 2 |
Hb_030736_070 |
0.0856825048 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000139_130 |
0.0897335558 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 4 |
Hb_000457_140 |
0.0940577787 |
- |
- |
PREDICTED: transmembrane protein 136-like [Jatropha curcas] |
| 5 |
Hb_000359_140 |
0.1058291209 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 6 |
Hb_146081_010 |
0.107161748 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 7 |
Hb_008143_070 |
0.1094277035 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 8 |
Hb_001258_110 |
0.110845026 |
- |
- |
transporter, putative [Ricinus communis] |
| 9 |
Hb_004531_120 |
0.1143883707 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 10 |
Hb_001486_070 |
0.1156774908 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 11 |
Hb_015026_130 |
0.1173228137 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 12 |
Hb_001033_050 |
0.1179434259 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 13 |
Hb_002662_020 |
0.118492581 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 14 |
Hb_000692_150 |
0.1193759149 |
- |
- |
PREDICTED: probable prolyl 4-hydroxylase 3 [Jatropha curcas] |
| 15 |
Hb_004109_230 |
0.1212036038 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 16 |
Hb_004668_060 |
0.1215030226 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 17 |
Hb_131035_010 |
0.121735314 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_001205_110 |
0.122574821 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 19 |
Hb_006573_210 |
0.1238622217 |
- |
- |
PREDICTED: protein SGT1 homolog [Jatropha curcas] |
| 20 |
Hb_002983_080 |
0.1239568337 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |