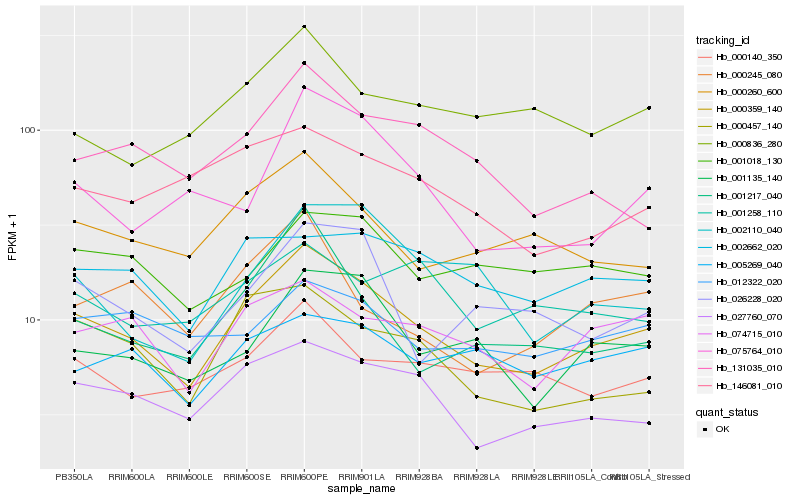
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000359_140 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 2 |
Hb_027760_070 |
0.1058291209 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 3 |
Hb_001135_140 |
0.1208409763 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 4 |
Hb_074715_010 |
0.1213054054 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000260_600 |
0.1234365027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000457_140 |
0.1275087586 |
- |
- |
PREDICTED: transmembrane protein 136-like [Jatropha curcas] |
| 7 |
Hb_000140_350 |
0.1289167927 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_026228_020 |
0.1316326312 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 9 |
Hb_002662_020 |
0.1320148646 |
- |
- |
PREDICTED: F-box protein At1g47056-like [Jatropha curcas] |
| 10 |
Hb_131035_010 |
0.1323453679 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_146081_010 |
0.1372255372 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 12 |
Hb_001217_040 |
0.137295703 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 13 |
Hb_000836_280 |
0.1381217443 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005269_040 |
0.1388728483 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein At1g06270 [Jatropha curcas] |
| 15 |
Hb_000245_080 |
0.1401051777 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 16 |
Hb_002110_040 |
0.1402182843 |
- |
- |
hypothetical protein AMTR_s00135p00074190 [Amborella trichopoda] |
| 17 |
Hb_001018_130 |
0.1405923814 |
- |
- |
PREDICTED: uncharacterized protein LOC105638260 [Jatropha curcas] |
| 18 |
Hb_075764_010 |
0.1411161018 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 19 |
Hb_012322_020 |
0.143561875 |
- |
- |
- |
| 20 |
Hb_001258_110 |
0.1448439938 |
- |
- |
transporter, putative [Ricinus communis] |