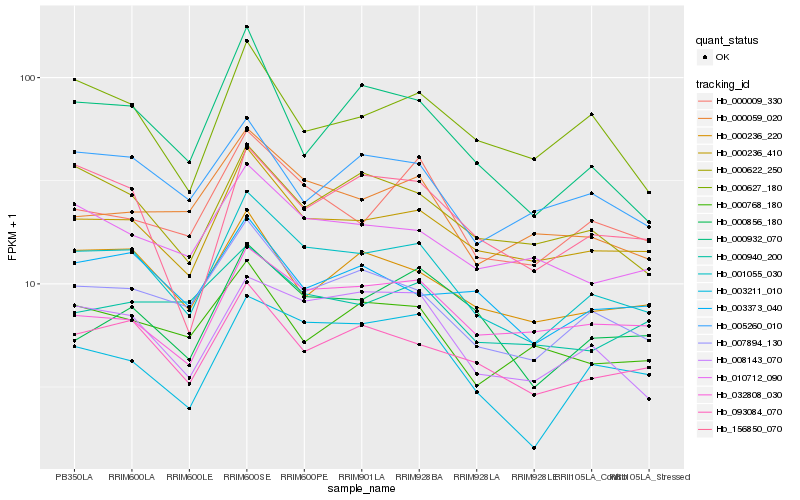
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001055_030 |
0.0 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007894_130 |
0.0723972045 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 3 |
Hb_000236_410 |
0.074970592 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 4 |
Hb_093084_070 |
0.0816381512 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 5 |
Hb_008143_070 |
0.0845970056 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 6 |
Hb_000009_330 |
0.0939782356 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_000622_250 |
0.0942421074 |
- |
- |
atpob1, putative [Ricinus communis] |
| 8 |
Hb_000236_220 |
0.0948712974 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 9 |
Hb_010712_090 |
0.0970063539 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 10 |
Hb_003211_010 |
0.0993941129 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 11 |
Hb_000059_020 |
0.1014816089 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 12 |
Hb_000627_180 |
0.1025404079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_032808_030 |
0.1025602225 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_003373_040 |
0.1039705277 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 15 |
Hb_005260_010 |
0.1062282767 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_156850_070 |
0.1068644425 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000932_070 |
0.1079963789 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 18 |
Hb_000856_180 |
0.1100332435 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 19 |
Hb_000768_180 |
0.1100815839 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 20 |
Hb_000940_200 |
0.1112922097 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |