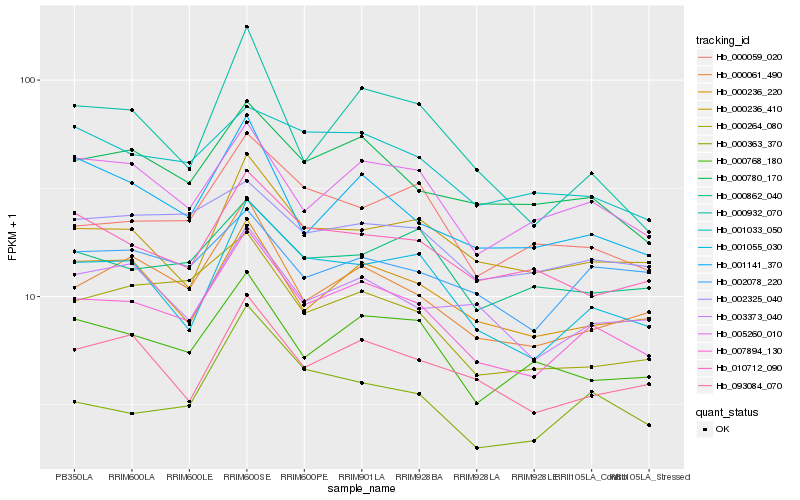
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_130 |
0.0 |
- |
- |
hypothetical protein RCOM_0584960 [Ricinus communis] |
| 2 |
Hb_001055_030 |
0.0723972045 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 14 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000061_490 |
0.0746541407 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 4 |
Hb_000780_170 |
0.0786942853 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 5 |
Hb_005260_010 |
0.0837844188 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000768_180 |
0.0858535696 |
- |
- |
PREDICTED: uncharacterized protein LOC105632528 [Jatropha curcas] |
| 7 |
Hb_000236_220 |
0.086356381 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-5 [Jatropha curcas] |
| 8 |
Hb_093084_070 |
0.086996332 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 9 |
Hb_000236_410 |
0.0887289083 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 10 |
Hb_000059_020 |
0.0915819928 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 11 |
Hb_001141_370 |
0.0917060693 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 12 |
Hb_003373_040 |
0.0922366352 |
- |
- |
hypothetical protein ARALYDRAFT_356474 [Arabidopsis lyrata subsp. lyrata] |
| 13 |
Hb_000264_080 |
0.0931100169 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 14 |
Hb_010712_090 |
0.0933765139 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 15 |
Hb_000363_370 |
0.0957013304 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 16 |
Hb_002325_040 |
0.0972546721 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001033_050 |
0.0980484747 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 18 |
Hb_000932_070 |
0.0983963182 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 19 |
Hb_002078_220 |
0.0989881065 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000862_040 |
0.0992614893 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |