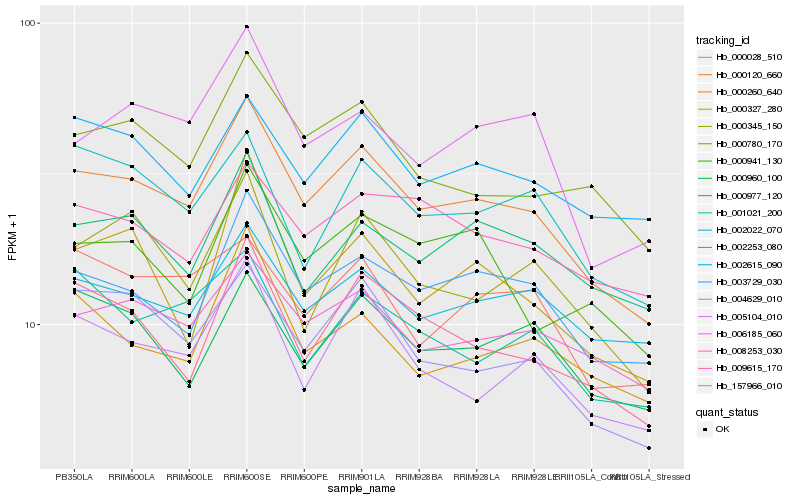
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_660 |
0.0 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 2 |
Hb_003729_030 |
0.064709185 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 3 |
Hb_004629_010 |
0.0663593004 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 4 |
Hb_157966_010 |
0.0704181666 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 5 |
Hb_005104_010 |
0.0710042197 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 6 |
Hb_000780_170 |
0.0719290301 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |
| 7 |
Hb_008253_030 |
0.0744599918 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 8 |
Hb_006185_060 |
0.0760644874 |
- |
- |
PREDICTED: protein phosphatase 2C 16 [Jatropha curcas] |
| 9 |
Hb_000345_150 |
0.0803282251 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002615_090 |
0.0807558709 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000028_510 |
0.082560708 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 12 |
Hb_000327_280 |
0.0827690915 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000260_640 |
0.0827719482 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 14 |
Hb_002253_080 |
0.0832112763 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 15 |
Hb_000977_120 |
0.0837049728 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 4 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000960_100 |
0.0839745173 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 17 |
Hb_002022_070 |
0.0842787433 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_000941_130 |
0.0844250884 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 19 |
Hb_009615_170 |
0.0846437826 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001021_200 |
0.085661239 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |