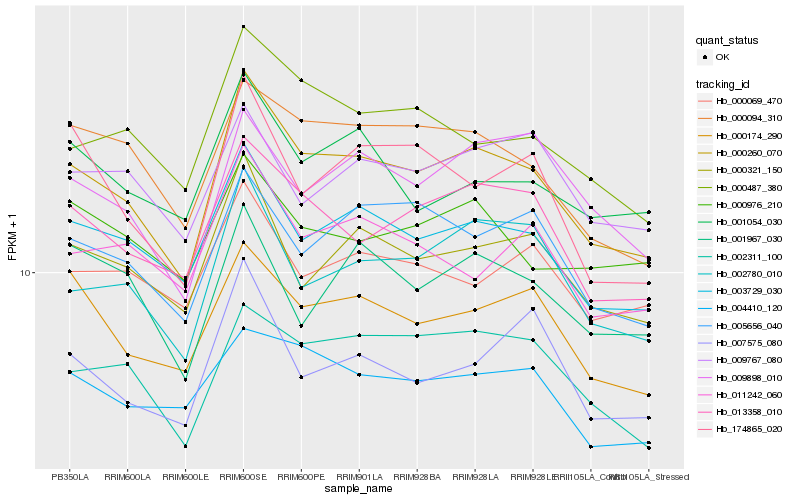
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_070 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_003729_030 |
0.0648472197 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 3 |
Hb_013358_010 |
0.0732958258 |
- |
- |
hypothetical protein JCGZ_07102 [Jatropha curcas] |
| 4 |
Hb_000321_150 |
0.0776367055 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002780_010 |
0.0802224123 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 6 |
Hb_000174_290 |
0.0809461006 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 7 |
Hb_000094_310 |
0.081182035 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 8 |
Hb_002311_100 |
0.0823498666 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 9 |
Hb_000487_380 |
0.083521719 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005656_040 |
0.0866854546 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 11 |
Hb_009898_010 |
0.0904087312 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 12 |
Hb_011242_060 |
0.0928264774 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 13 |
Hb_004410_120 |
0.0934079473 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 14 |
Hb_174865_020 |
0.0943221713 |
- |
- |
E3 ubiquitin protein ligase upl2, putative [Ricinus communis] |
| 15 |
Hb_001054_030 |
0.09442617 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 16 |
Hb_000069_470 |
0.0968567295 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007575_080 |
0.0976656331 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 18 |
Hb_001967_030 |
0.098710865 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 19 |
Hb_009767_080 |
0.099951385 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 20 |
Hb_000976_210 |
0.1003161017 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |