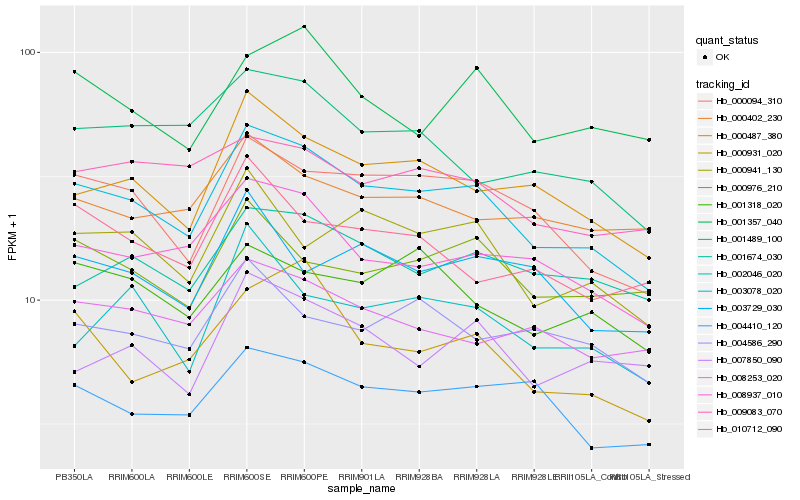
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003078_020 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 2 |
Hb_000094_310 |
0.0724579289 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g75140 [Jatropha curcas] |
| 3 |
Hb_000487_380 |
0.0760837802 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_008937_010 |
0.0808802572 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80A2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000941_130 |
0.0866738559 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 6 |
Hb_004410_120 |
0.0917808001 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001489_100 |
0.0924375926 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 8 |
Hb_001674_030 |
0.0941996472 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g24240 [Jatropha curcas] |
| 9 |
Hb_000931_020 |
0.0963744814 |
- |
- |
PREDICTED: uncharacterized protein LOC105649774 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008253_020 |
0.0966801042 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 11 |
Hb_001318_020 |
0.0982534145 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 12 |
Hb_001357_040 |
0.0987040128 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 13 |
Hb_009083_070 |
0.0991087179 |
- |
- |
PREDICTED: uncharacterized protein LOC105639870 [Jatropha curcas] |
| 14 |
Hb_002046_020 |
0.0998425372 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 15 |
Hb_000402_230 |
0.1003287914 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000976_210 |
0.100550544 |
- |
- |
PREDICTED: protein PAT1 homolog 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_010712_090 |
0.1008378922 |
- |
- |
PREDICTED: uncharacterized protein LOC105631276 [Jatropha curcas] |
| 18 |
Hb_003729_030 |
0.1011104597 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 19 |
Hb_007850_090 |
0.1011926969 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004586_290 |
0.1013783449 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |